Search Count: 58
 |
Organism: Glycine max
Method: X-RAY DIFFRACTION Release Date: 2025-06-04 Classification: PLANT PROTEIN Ligands: CYC |
 |
Organism: Oscillatoria acuminata
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2025-02-26 Classification: LYASE Ligands: FMN, GOL, CA |
 |
Organism: Glycine max
Method: X-RAY DIFFRACTION Release Date: 2025-01-08 Classification: PLANT PROTEIN Ligands: CYC, 1PE, PEG, PGE |
 |
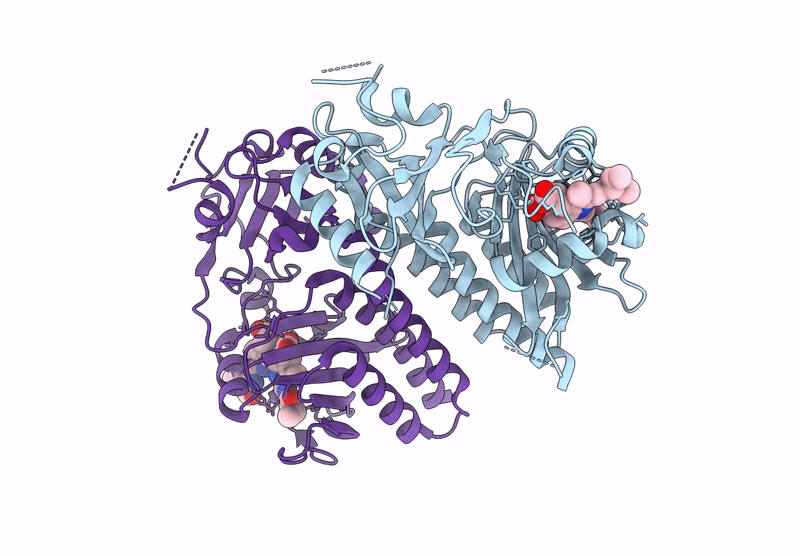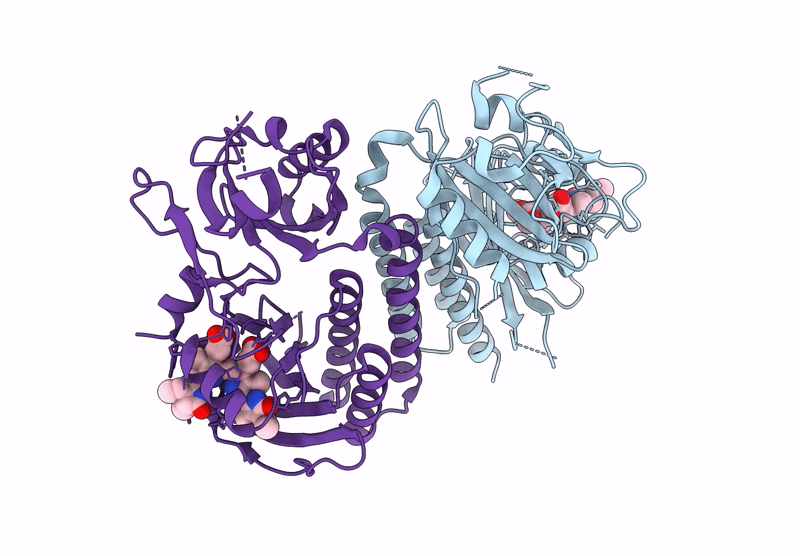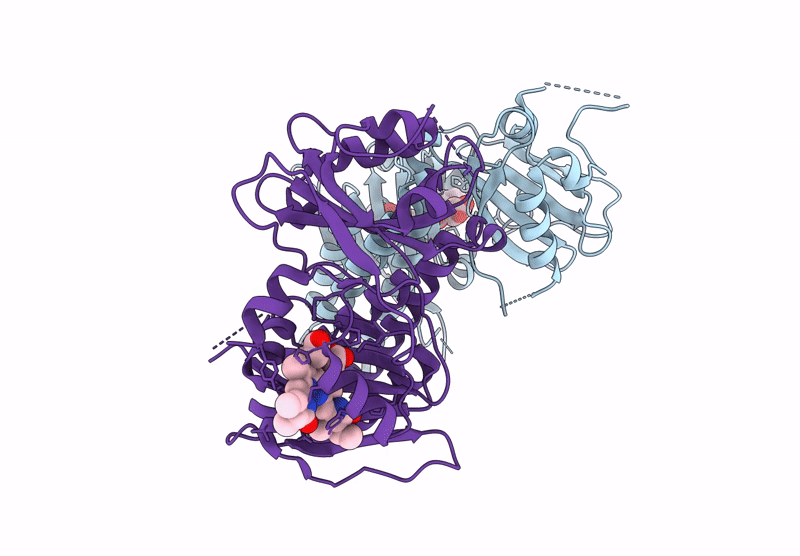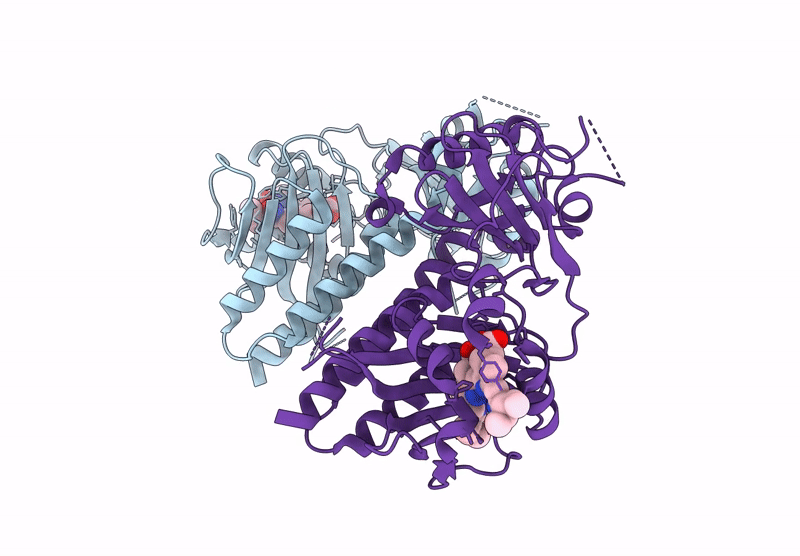
Organism: Glycine max
Method: X-RAY DIFFRACTION Release Date: 2025-01-08 Classification: PLANT PROTEIN Ligands: O6E, PG4 |
 |
Organism: Glycine max
Method: X-RAY DIFFRACTION Release Date: 2025-01-08 Classification: PLANT PROTEIN Ligands: CYC |
 |
Organism: Glycine max
Method: X-RAY DIFFRACTION Release Date: 2025-01-08 Classification: PLANT PROTEIN Ligands: CYC |
 |
Room Temperature Structure Of Fad-Containing Ferrodoxin-Nadp Reductase From Brucella Ovis At Euxfel
Organism: Brucella ovis atcc 25840
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-11-27 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Room Temperature Structure Of Fad-Containing Ferrodoxin-Nadp Reductase From Brucella Ovis At Lcls
Organism: Brucella ovis atcc 25840
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-11-27 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Hen Egg-White Lysozyme (Hewl) Structure From Euxfel Fxe, Multi-Hit Droplet-On-Demand (Dod) Injection, 9.3 Kev Photon Energy, Space Group P432121
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2024-07-24 Classification: HYDROLASE Ligands: CL, NA, ACT, PEG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-06-26 Classification: TRANSFERASE/INHIBITOR Ligands: UEO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-06-26 Classification: TRANSFERASE/INHIBITOR Ligands: V72 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-06-26 Classification: TRANSFERASE/INHIBITOR Ligands: V7I |
 |
Crystal Structure Of The Main Protease (3Clpro/Mpro) Of Sars-Cov-2 Obtained In Presence Of 150 Micromolar Mg-132.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-05-01 Classification: VIRAL PROTEIN Ligands: ALD, CL, NA, DMS, EDO |
 |
Crystal Structure Of The Main Protease (3Clpro/Mpro) Of Sars-Cov-2 Obtained In Presence Of 75 Micromolar Mg-132.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-05-01 Classification: VIRAL PROTEIN |
 |
Crystal Structure Of The Main Protease (3Clpro/Mpro) Of Sars-Cov-2 Obtained In Presence Of 150 Micromolar X77.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2024-05-01 Classification: VIRAL PROTEIN Ligands: EDO, CL, X77, DMS, NA |
 |
Crystal Structure Of The Main Protease (3Clpro/Mpro) Of Sars-Cov-2 Obtained In Presence Of 75 Micromolar X77.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-05-01 Classification: VIRAL PROTEIN Ligands: CL, X77, EDO, DMS, NA |
 |
Crystal Structure Of The Main Protease (3Clpro/Mpro) Of Sars-Cov-2 Obtained In Presence Of 500 Micromolar X77 Enantiomer R.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2024-05-01 Classification: VIRAL PROTEIN Ligands: EDO, CL, X77, DMS, NA |
 |
Crystal Structure Of The Main Protease (3Clpro/Mpro) Of Sars-Cov-2 Obtained In Presence Of 5 Millimolar X77 Enantiomer R.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2024-05-01 Classification: VIRAL PROTEIN Ligands: EDO, CL, X77, DMS, NA |
 |
Crystal Structure Of The Main Protease (3Clpro/Mpro) Of Sars-Cov-2 Obtained In Presence Of 500 Micromolar X77 Enantiomer S.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2024-05-01 Classification: VIRAL PROTEIN Ligands: DMS, EDO, CL, NA |
 |
Crystal Structure Of The Main Protease (3Clpro/Mpro) Of Sars-Cov-2 Obtained In Presence Of 5 Millimolar X77 Enantiomer S.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2024-05-01 Classification: VIRAL PROTEIN Ligands: 9M5, EDO, ACT, DMS, CL, NA |

