Search Count: 92
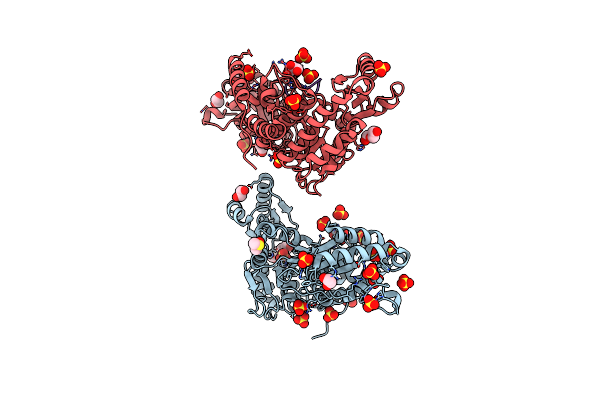 |
Mycobacterium Tuberculosis Pks13 Acyltransferase Serine Converted To Beta-Lactam Form By Cec215 Via Sufex Reaction
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Release Date: 2025-05-21 Classification: ANTIBIOTIC Ligands: SO4, CL, PG4, EDO, DMS, GOL, PEG |
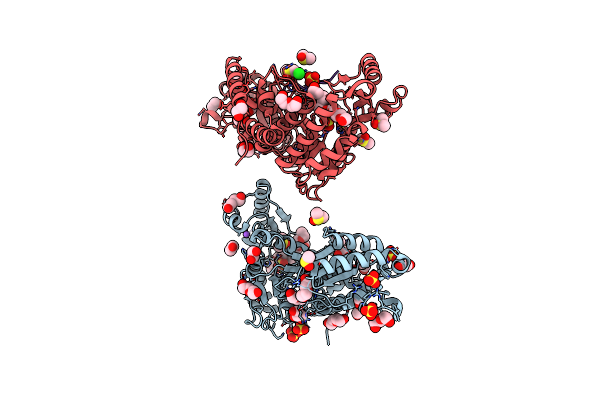 |
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Release Date: 2025-05-21 Classification: ANTIBIOTIC Ligands: DMS, SO4, PE5, EDO, PEG, GOL, CL, NA, P33 |
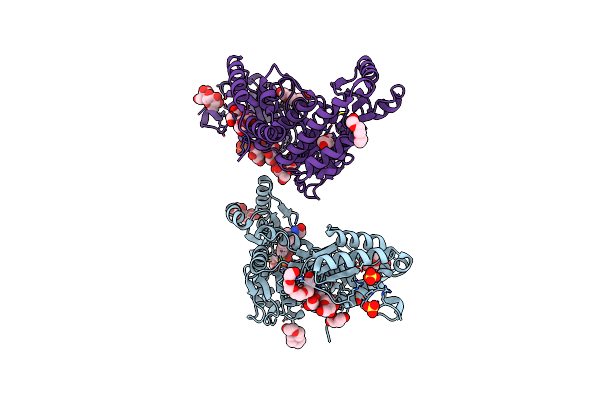 |
M. Tuberculosis Pks13 Acyltransferase (At) Domain In Complex With Sufex Inhibitor Cec215
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Release Date: 2025-05-07 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 1PE, SO4, A1ATV |
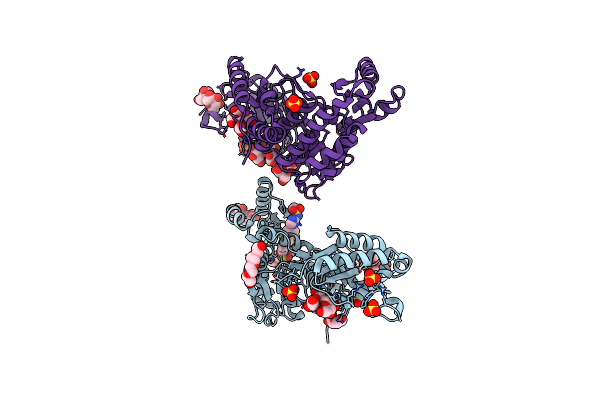 |
M. Tuberculosis Pks13 Acyltransferase (At) Domain In Complex With Sufex Inhibitor Cmx410
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Release Date: 2025-05-07 Classification: TRANSFERASE/TRANSFERSE INHIBITOR Ligands: 1PE, SO4, A1ATW |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Release Date: 2025-05-07 Classification: TRANSFERASE Ligands: 1PE, SO4 |
 |
M. Tuberculosis Pks13 Acyltransferase (At) Domain In Complex With Sufex Inhibitor Cmx410 - Reaction Product
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Release Date: 2025-05-07 Classification: TRANSFERASE Ligands: SO4, 1PE, A1AVL |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-08-07 Classification: TRANSCRIPTION |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-08-07 Classification: TRANSCRIPTION |
 |
Organism: Xenopus laevis, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-08-09 Classification: GENE REGULATION Ligands: ZN |
 |
Organism: Komagataella phaffii gs115
Method: ELECTRON MICROSCOPY Release Date: 2023-05-31 Classification: TRANSCRIPTION Ligands: ATP, MG |
 |
An Orthogonal Seryl-Trna Synthetase/Trna Pair For Noncanonical Amino Acid Mutagenesis In Escherichia Coli
Organism: Methanosarcina mazei (strain atcc baa-159 / dsm 3647 / goe1 / go1 / jcm 11833 / ocm 88)
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2020-11-04 Classification: LIGASE Ligands: SSA, PEG, DMS, SCN, K, CL |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2020-08-26 Classification: IMMUNE SYSTEM Ligands: V67 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2020-08-26 Classification: IMMUNE SYSTEM Ligands: V67, GOL, EDO |
 |
Organism: Komagataella phaffii (strain gs115 / atcc 20864), Homo sapiens, Komagataella phaffii gs115
Method: ELECTRON MICROSCOPY Release Date: 2020-02-12 Classification: TRANSCRIPTION |
 |
Organism: Komagataella phaffii (strain gs115 / atcc 20864), Komagataella phaffii gs115
Method: ELECTRON MICROSCOPY Release Date: 2020-01-29 Classification: TRANSCRIPTION |
 |
A Single Reactive Noncanonical Amino Acid Is Able To Dramatically Stabilize Protein Structure
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2019-06-26 Classification: TRANSFERASE Ligands: PEG, FMT, GOL |
 |
Organism: Komagataella phaffii gs115, Komagataella phaffii (strain gs115 / atcc 20864)
Method: ELECTRON MICROSCOPY Release Date: 2018-11-21 Classification: TRANSCRIPTION |
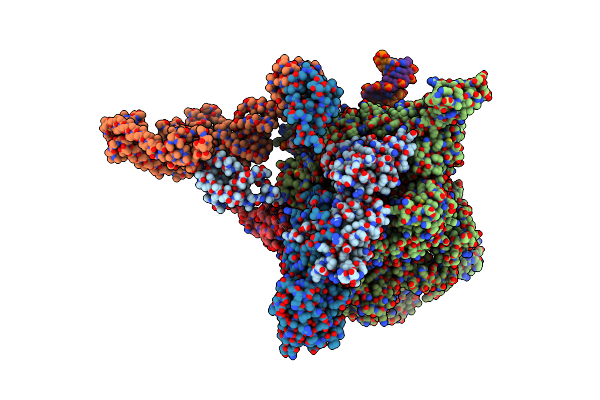 |
Cryoem Structure Of E.Coli Rna Polymerase Paused Elongation Complex Bound To Nusa
Organism: Escherichia coli (strain k12), Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2018-03-21 Classification: TRANSCRIPTION Ligands: MG, ZN |
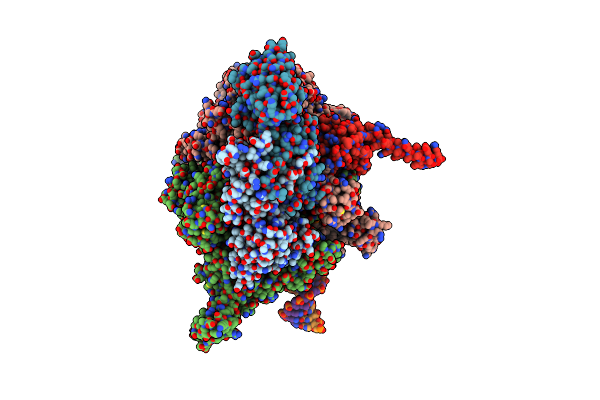 |
Cryoem Structure Of E.Coli Rna Polymerase Paused Elongation Complex Without Rna Hairpin Bound To Nusa
Organism: Escherichia coli (strain k12), Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2018-03-07 Classification: TRANSCRIPTION Ligands: MG, ZN |
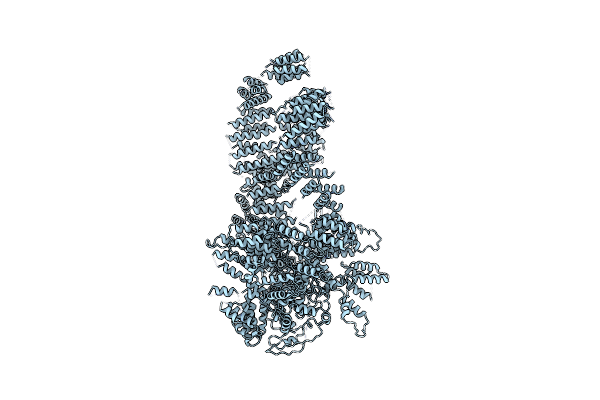 |
Organism: Komagataella pastoris
Method: ELECTRON MICROSCOPY Resolution:5.70 Å Release Date: 2017-08-02 Classification: TRANSCRIPTION |

