Search Count: 23
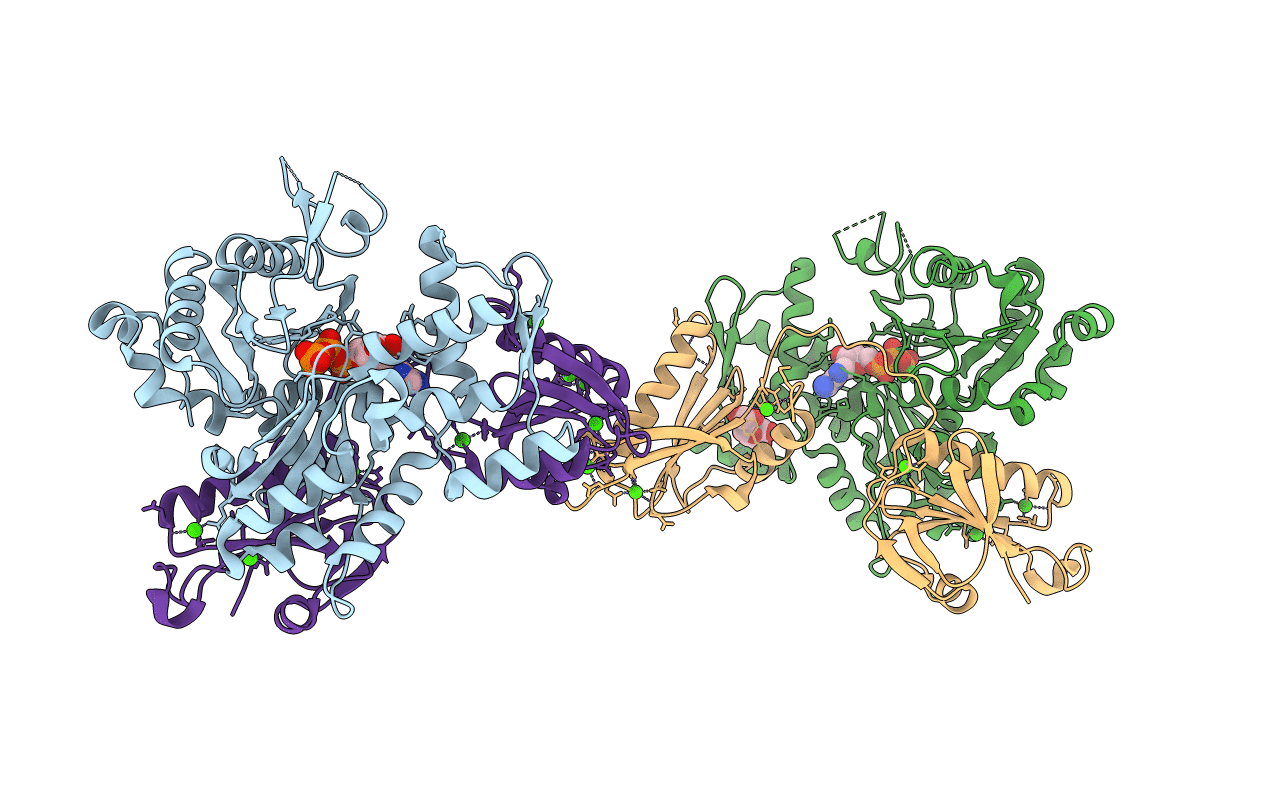 |
Organism: Oryctolagus cuniculus, Candidatus heimdallarchaeota archaeon lc_3
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-09-21 Classification: STRUCTURAL PROTEIN Ligands: ATP, CA, GOL |
 |
Organism: Oryctolagus cuniculus, Candidatus lokiarchaeota archaeon
Method: X-RAY DIFFRACTION Resolution:3.25 Å Release Date: 2022-09-21 Classification: STRUCTURAL PROTEIN Ligands: ADP, CA |
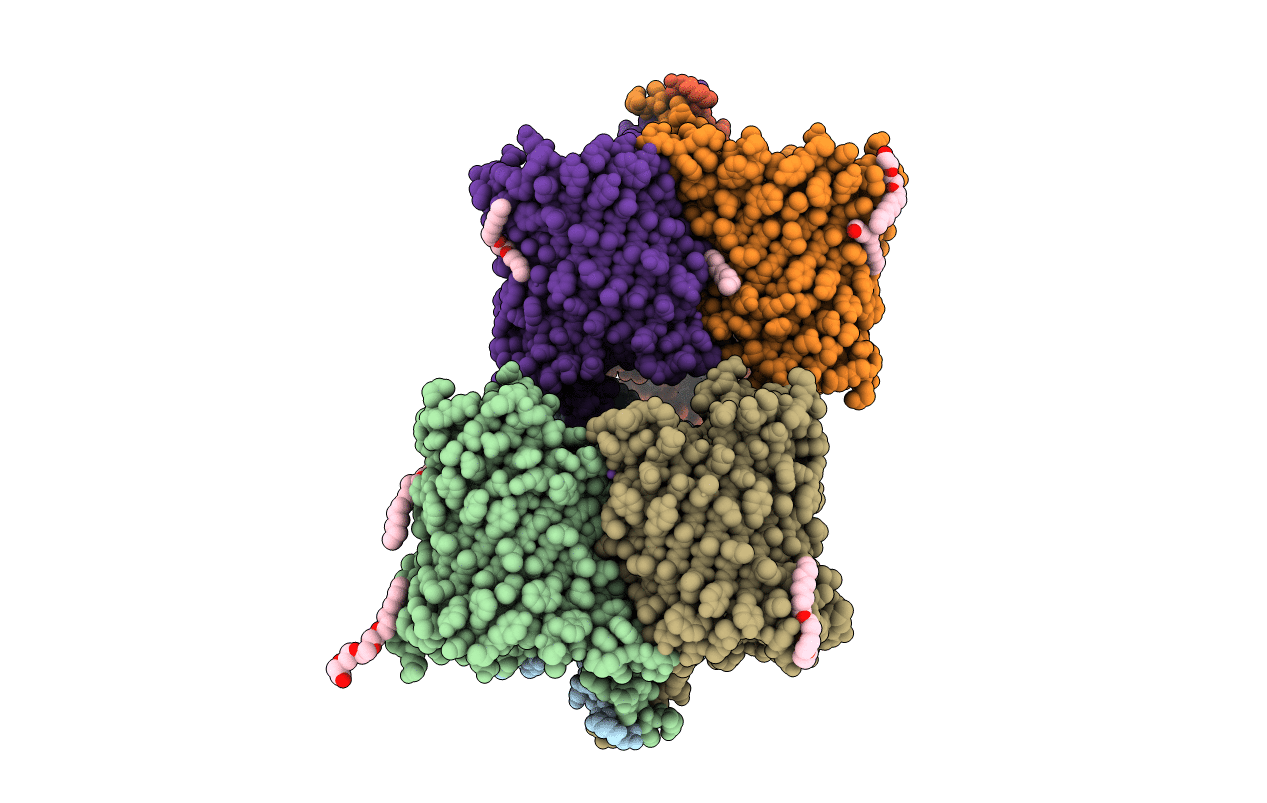 |
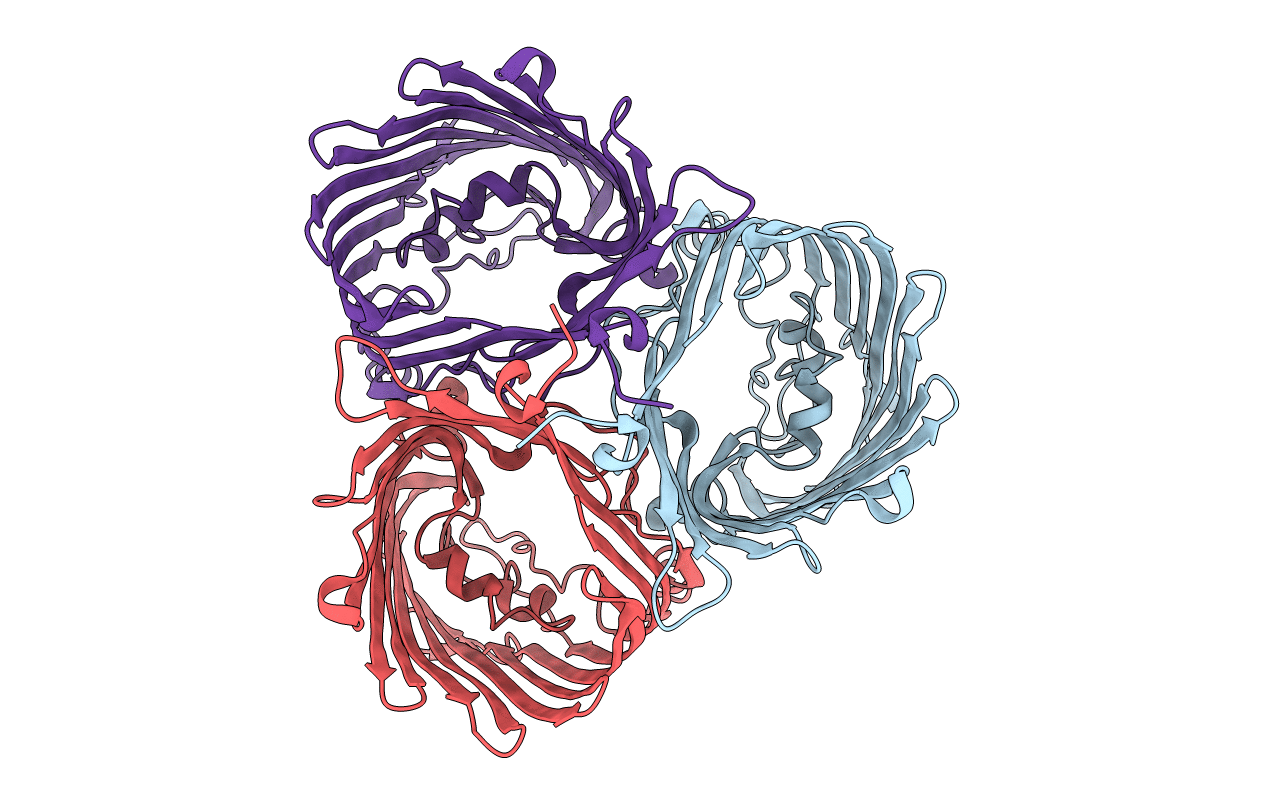
Crystal Structure Of In Vitro Folded Chitoporin Vhchip From Vibrio Harveyi (Crystal Form I)
Organism: Vibrio harveyi
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2017-12-20 Classification: SUGAR BINDING PROTEIN |
 |
Crystal Structure Of In Vitro Folded Chitoporin Vhchip From Vibrio Harveyi (Crystal Form Ii)
Organism: Vibrio harveyi
Method: X-RAY DIFFRACTION Resolution:3.08 Å Release Date: 2017-12-20 Classification: SUGAR BINDING PROTEIN |
 |
Crystal Structure Of Outer Membrane Expressed Chitoporin Vhchip From Vibrio Harveyi
Organism: Vibrio harveyi
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2017-12-20 Classification: SUGAR BINDING PROTEIN |
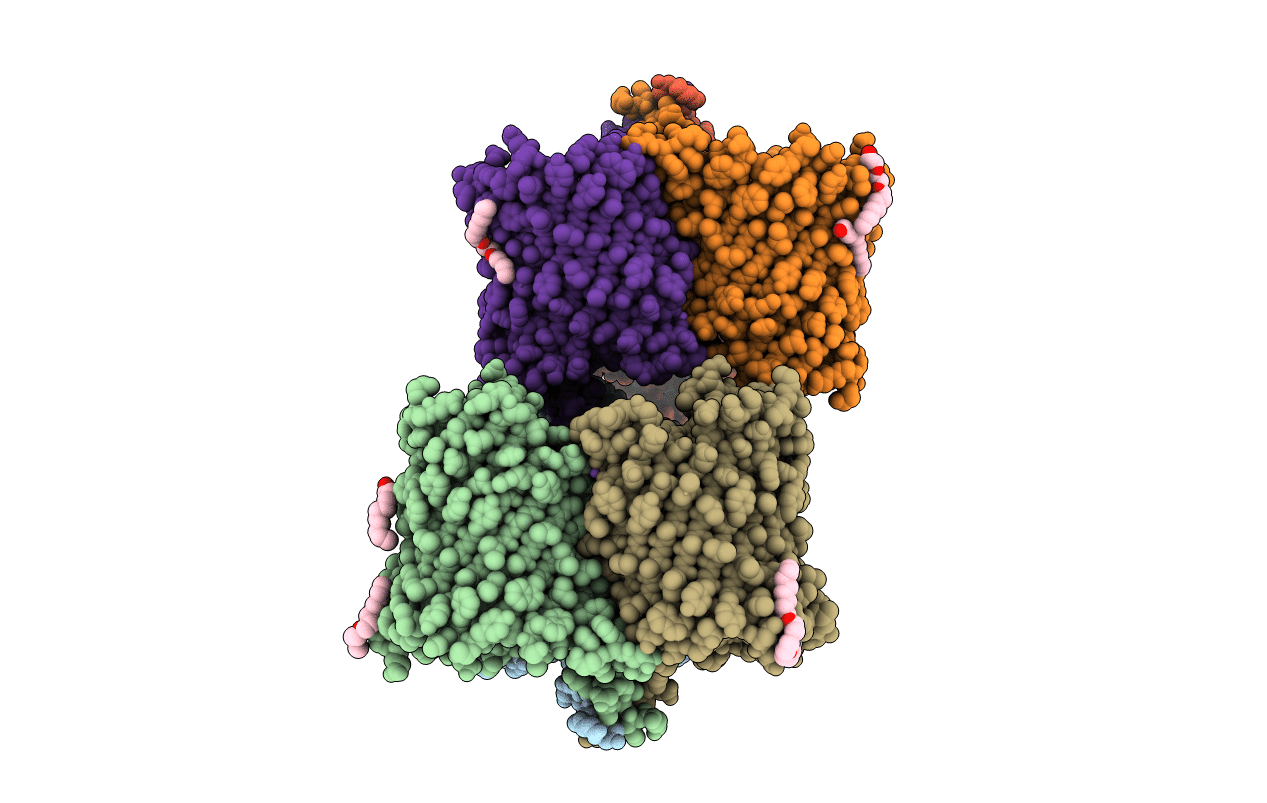 |
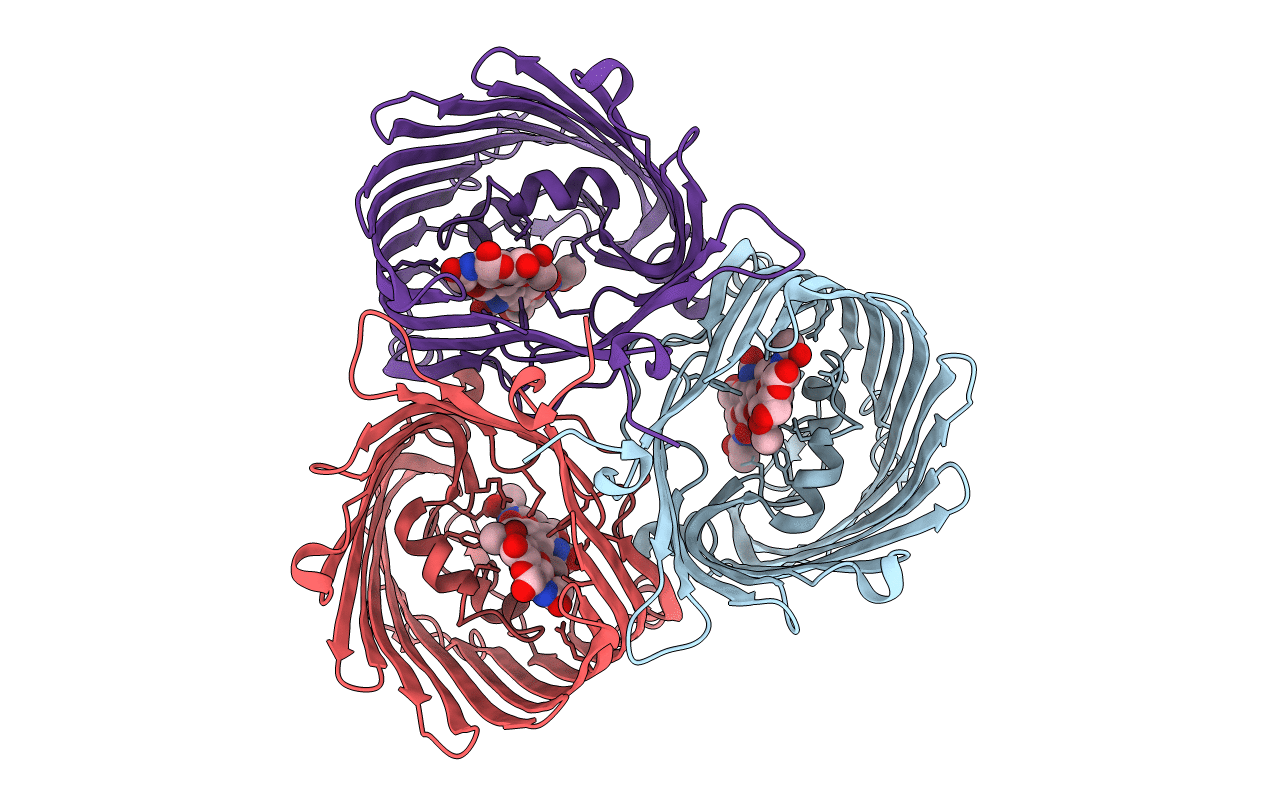
Crystal Structure Of In Vitro Folded Chitoporin Vhchip From Vibrio Harveyi In Complex With Chitohexaose
Organism: Vibrio harveyi
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2017-12-20 Classification: SUGAR BINDING PROTEIN Ligands: C8E, NA |
 |
Crystal Structure Of Outer Membrane Expressed Chitoporin Vhchip From Vibrio Harveyi In Complex With Chitotetraose
Organism: Vibrio harveyi
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2017-12-20 Classification: SUGAR BINDING PROTEIN |
 |
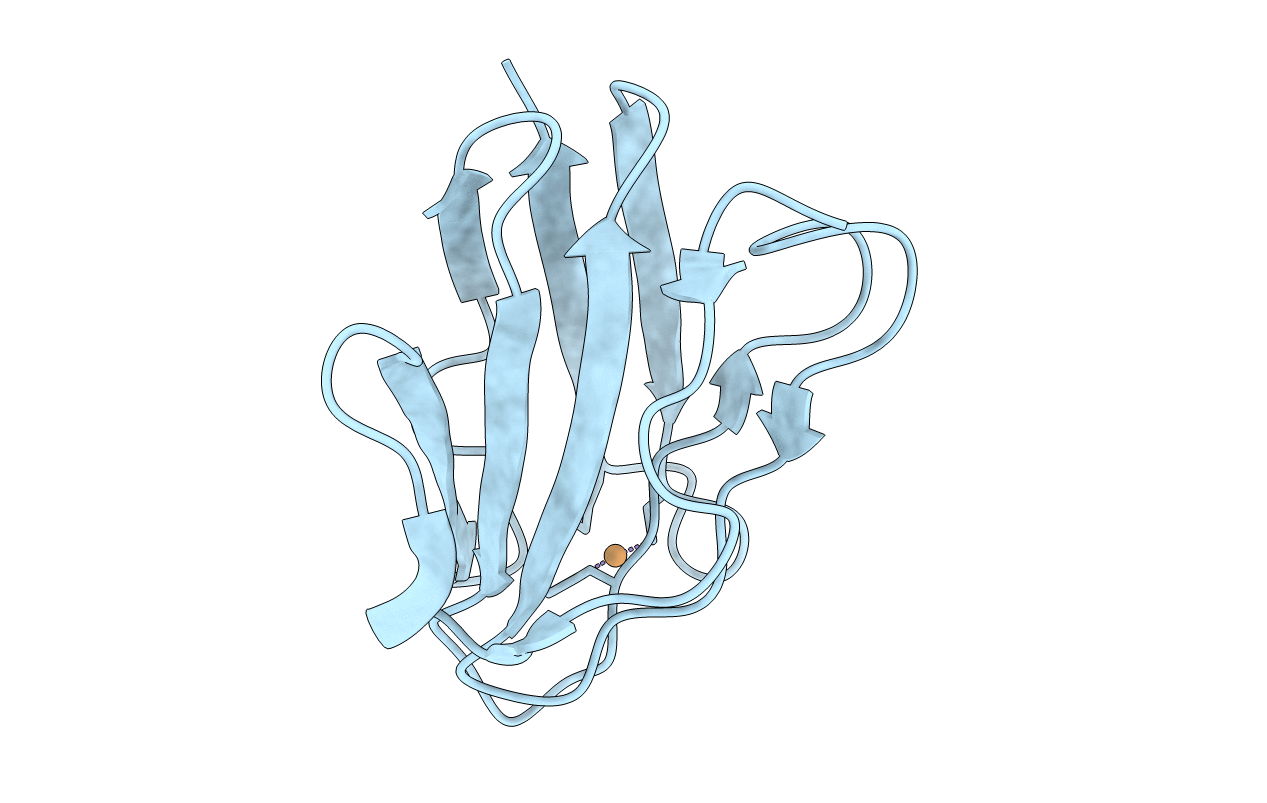
Organism: Paracoccus denitrificans
Method: X-RAY DIFFRACTION Resolution:1.09 Å Release Date: 2014-04-23 Classification: ELECTRON TRANSPORT Ligands: CU, NA |
 |
Organism: Paracoccus denitrificans
Method: X-RAY DIFFRACTION Resolution:1.02 Å Release Date: 2014-04-23 Classification: ELECTRON TRANSPORT Ligands: CU1 |
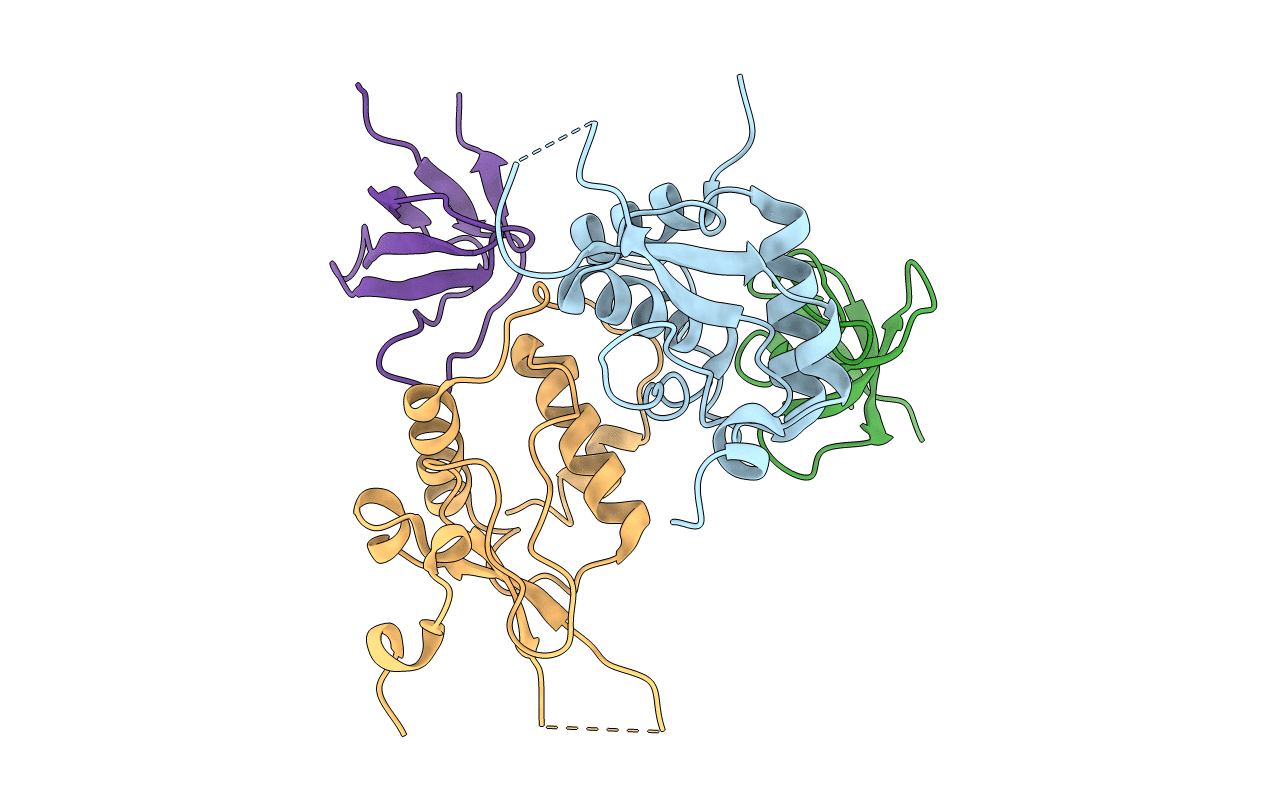 |
Organism: Hiv-1 m:b_arv2/sf2, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-06-01 Classification: PROTEIN BINDING |
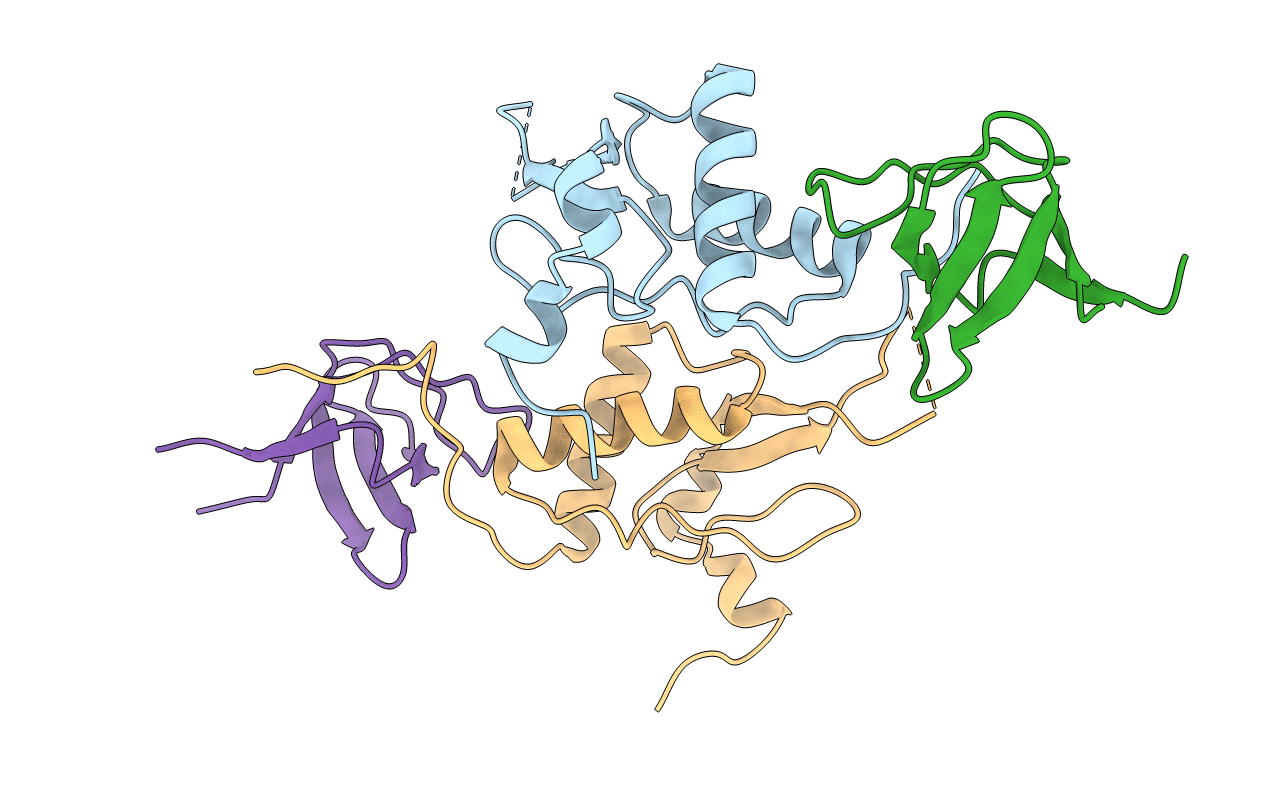 |
Organism: Hiv-1 m:b_arv2/sf2, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.45 Å Release Date: 2011-06-01 Classification: PROTEIN BINDING |
 |
Organism: Hiv-1 m:b_arv2/sf2, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2011-04-27 Classification: VIRAL PROTEIN, PROTEIN BINDING Ligands: EDO |
 |
Crystal Structure Of Xanthine Dehydrogenase (Desulfo Form) From Rhodobacter Capsulatus In Complex With Hypoxanthine
Organism: Rhodobacter capsulatus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2008-12-23 Classification: OXIDOREDUCTASE Ligands: FES, FAD, MTE, CA, HPA, MOM |
 |
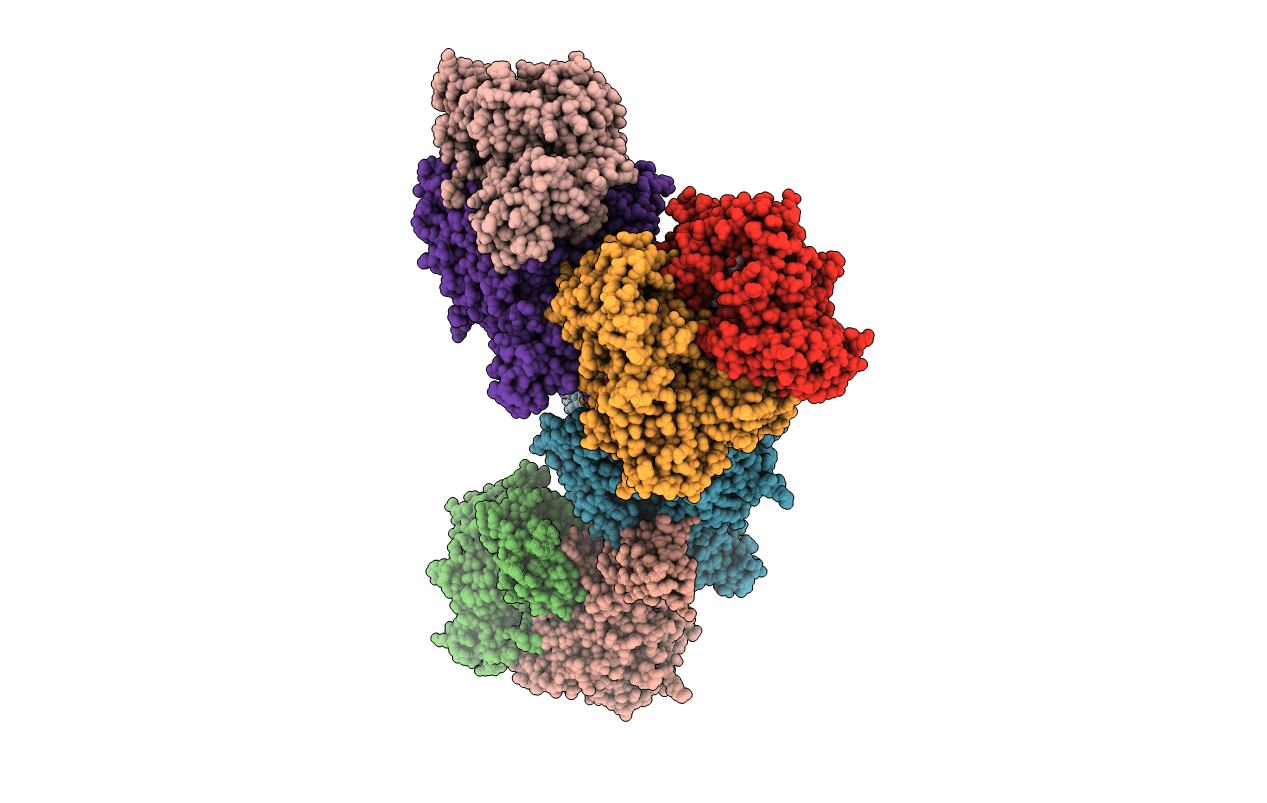
Crystal Structure Of Xanthine Dehydrogenase (Desulfo Form) From Rhodobacter Capsulatus In Complex With Xanthine
Organism: Rhodobacter capsulatus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2008-12-23 Classification: OXIDOREDUCTASE Ligands: FES, FAD, MTE, CA, XAN, MOM |
 |
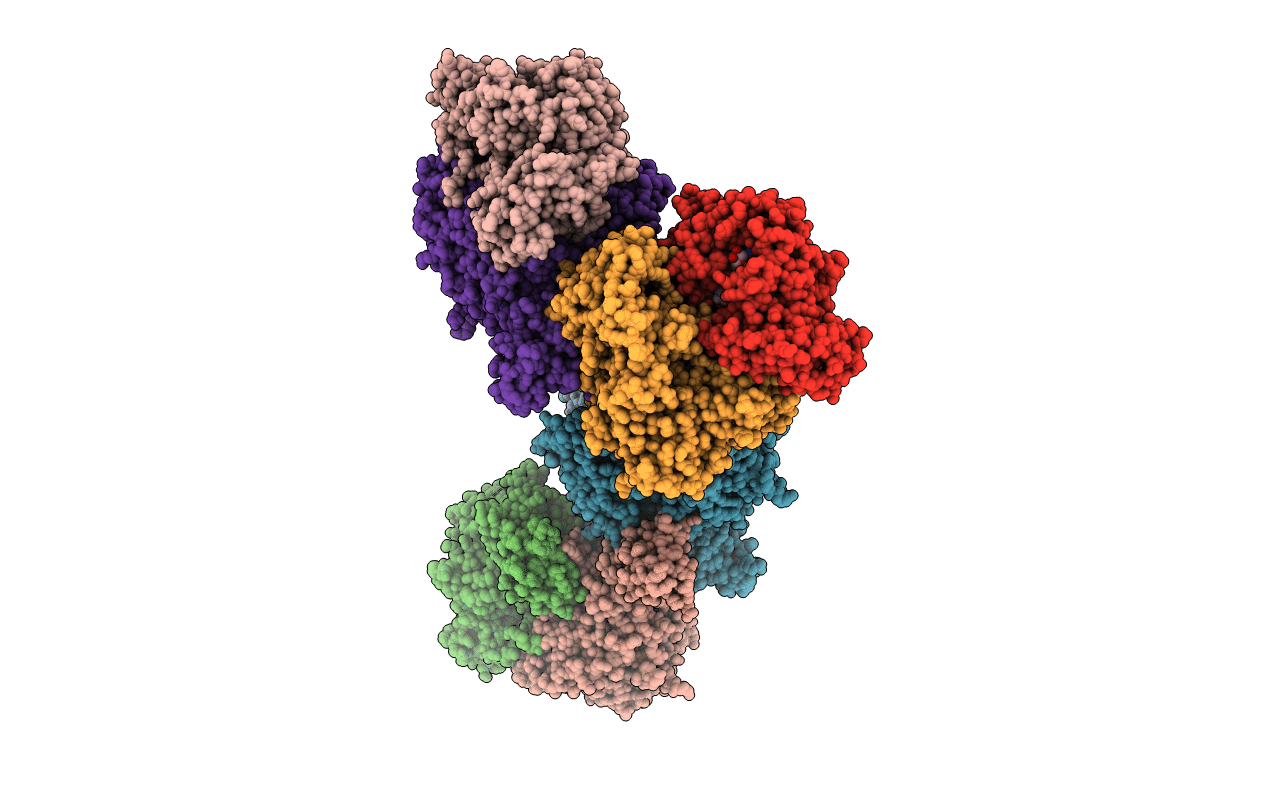
Crystal Structure Of Xanthine Dehydrogenase From Rhodobacter Capsulatus In Complex With Bound Inhibitor Pterin-6-Aldehyde
Organism: Rhodobacter capsulatus
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2008-12-23 Classification: OXIDOREDUCTASE Ligands: FES, FAD, XAX, BA, HHR |
 |
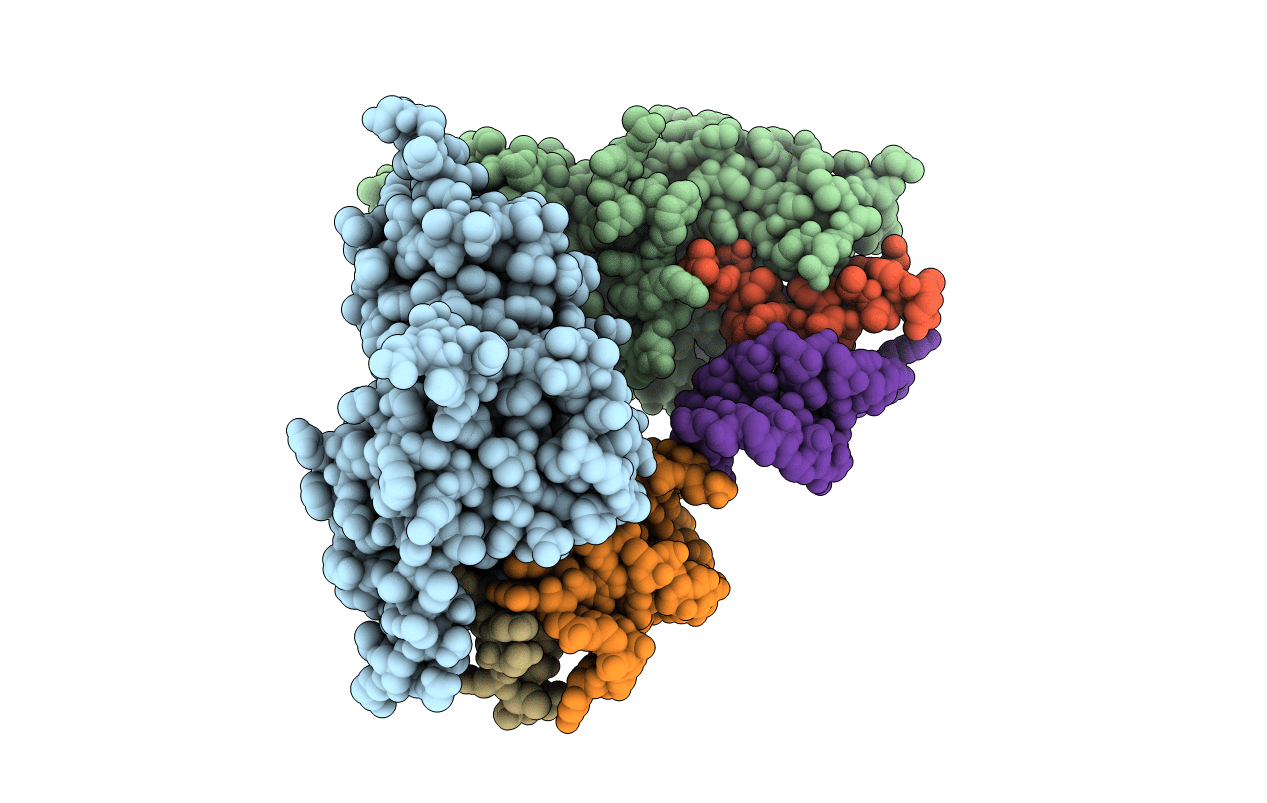
Crystal Structure Of Xanthine Dehydrogenase (E232Q Variant) From Rhodobacter Capsulatus In Complex With Hypoxanthine
Organism: Rhodobacter capsulatus
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2008-12-23 Classification: OXIDOREDUCTASE Ligands: FES, FAD, XAX, BA, HPA |
 |
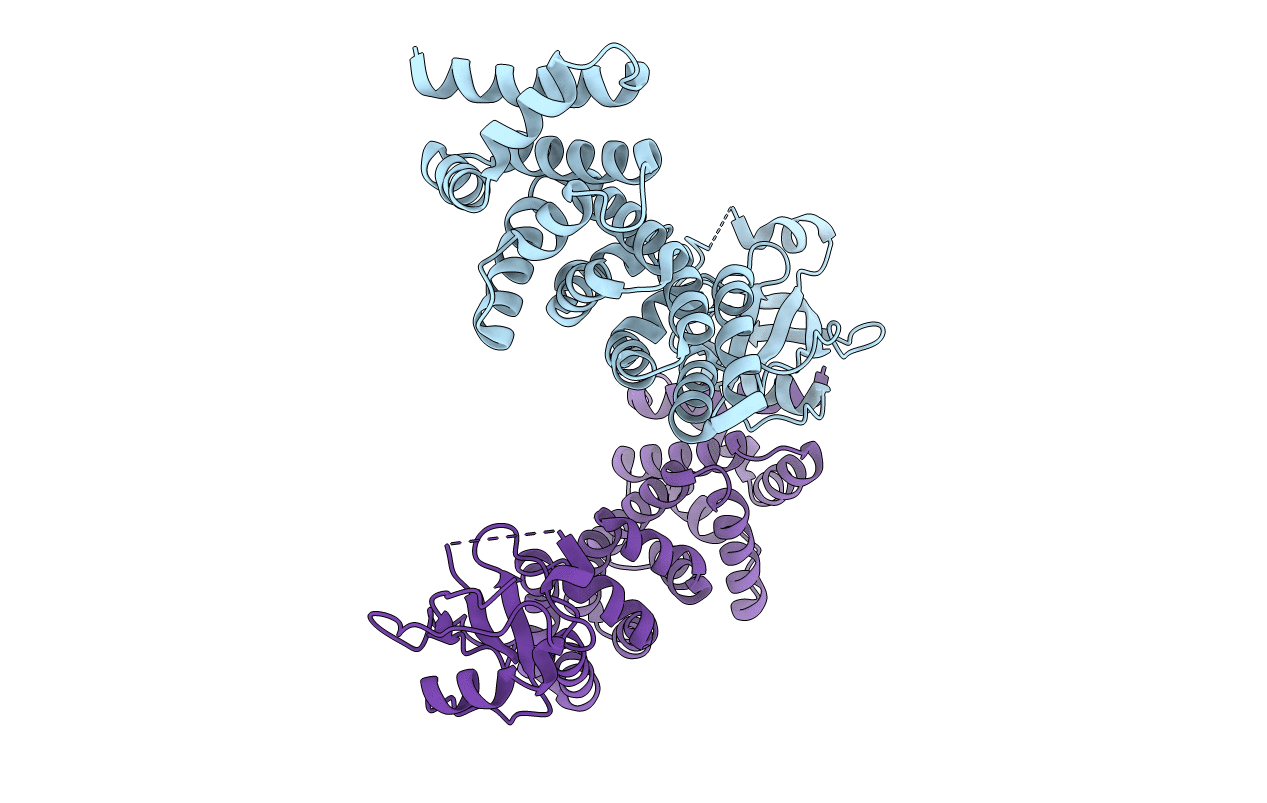
Structural Basis Of Transcription Activation By The Cyclin T1-Tat-Tar Rna Complex From Eiav
Organism: Equus caballus, Equine infectious anemia virus
Method: X-RAY DIFFRACTION Resolution:3.25 Å Release Date: 2008-12-09 Classification: RNA BINDING PROTEIN Ligands: MN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2008-09-16 Classification: SIGNALING PROTEIN |
 |
Organism: Allochromatium vinosum
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2007-07-03 Classification: TRANSFERASE |
 |
Cyclin Box Structure Of The P-Tefb Subunit Cyclin T1 Derived From A Fusion Complex With Eiav Tat
Organism: Homo sapiens, Equine infectious anemia virus
Method: X-RAY DIFFRACTION Resolution:2.67 Å Release Date: 2007-07-03 Classification: CELL CYCLE |

