Search Count: 37
 |
Crystal Structure Of 53Bp1 Tandem Tudor Domain Homodimer Engineered With Two Disulfide Bridges
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.79 Å Release Date: 2023-09-20 Classification: SIGNALING PROTEIN |
 |
Room Temperature Serial Crystal Structure Of Glutaminase C In Complex With Inhibitor Upgl-00004
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2022-05-25 Classification: HYDROLASE/INHIBITOR Ligands: B4A |
 |
Room Temperature Serial Crystal Structure Of Glutaminase C In Complex With Inhibitor Bptes
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2022-05-25 Classification: HYDROLASE/INHIBITOR Ligands: 04A |
 |
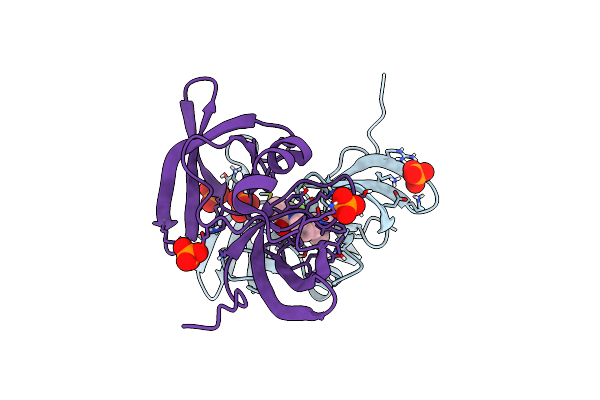
Structure Of 53Bp1 Tandem Tudor Domains In Complex With Small Molecule Unc2991
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2019-11-27 Classification: PROTEIN BINDING Ligands: PO4, K6P, FMT |
 |
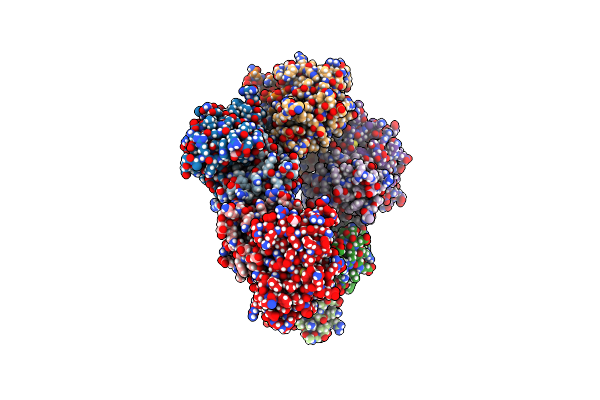
Structure Of 53Bp1 Tandem Tudor Domains In Complex With Small Molecule Unc3351
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2019-11-27 Classification: PROTEIN BINDING Ligands: PO4, K6M |
 |
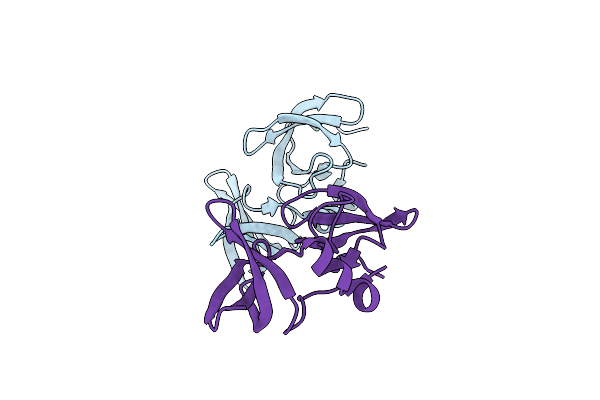
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2019-11-27 Classification: PROTEIN BINDING Ligands: K6S, FMT |
 |
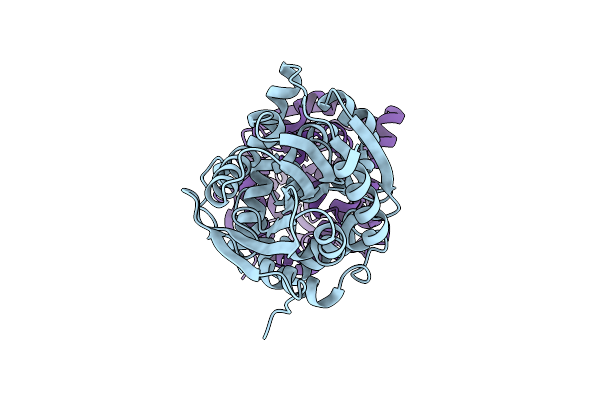
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2019-11-27 Classification: PROTEIN BINDING |
 |
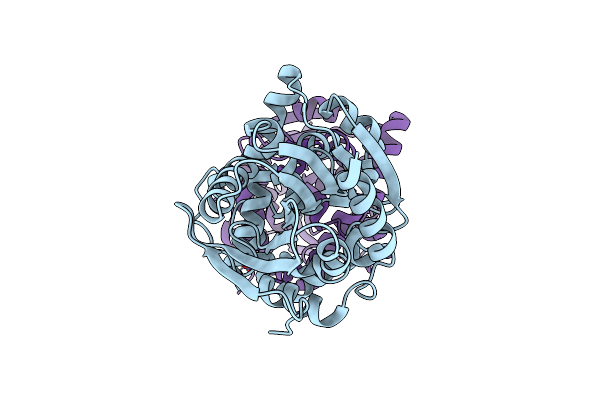
Fluoroacetate Dehalogenase, Room Temperature Structure Solved By Serial 1 Degree Oscillation Crystallography
Organism: Rhodopseudomonas palustris (strain atcc baa-98 / cga009)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-03-27 Classification: HYDROLASE |
 |
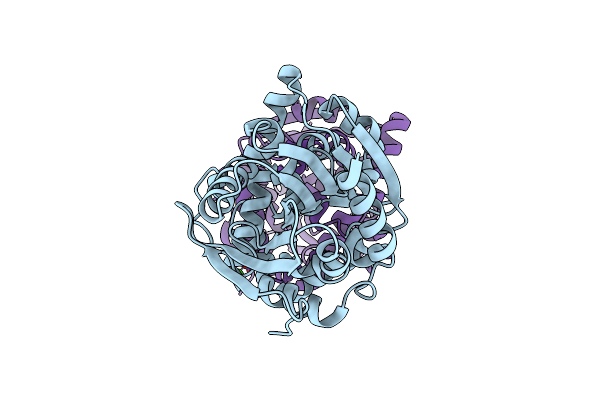
Fluoroacetate Dehalogenase, Room Temperature Structure Solved By Serial 3 Degree Oscillation Crystallography
Organism: Rhodopseudomonas palustris (strain atcc baa-98 / cga009)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-03-27 Classification: HYDROLASE Ligands: CA |
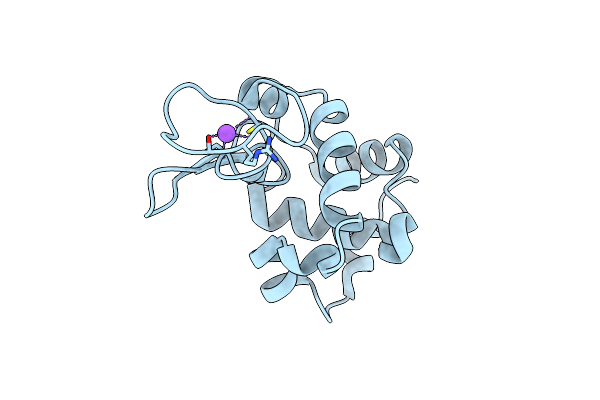 |
Lysozyme, Room Temperature Structure Solved By Serial 3 Degree Oscillation Crystallography
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2019-03-27 Classification: HYDROLASE Ligands: CL, NA |
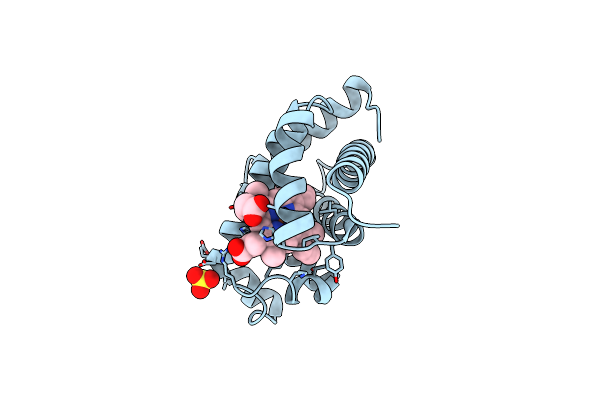 |
Co-Bound Sperm Whale Myoglobin, Room Temperature Structure Solved By Serial 5Degree Oscillation Crystallography
Organism: Physeter catodon
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2019-03-27 Classification: TRANSPORT PROTEIN Ligands: HEM, CMO, SO4 |
 |
Fluoroacetate Dehalogenase, Room Temperature Structure, Using First 1 Degree Of Total 3 Degree Oscillation
Organism: Rhodopseudomonas palustris cga009
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-03-27 Classification: HYDROLASE Ligands: CA |
 |
Fluoroacetate Dehalogenase, Room Temperature Structure, Using Last 1 Degree Of Total 3 Degree Oscillation And 144 Kgy Dose
Organism: Rhodopseudomonas palustris (strain atcc baa-98 / cga009)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-03-27 Classification: HYDROLASE Ligands: CA |
 |
Co-Bound Sperm Whale Myoglobin, Room Temperature Structure, First 2 Degrees Of 5 Degree Total Oscillation
Organism: Physeter catodon
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-03-27 Classification: OXYGEN TRANSPORT Ligands: HEM, CMO, SO4 |
 |
Co-Bound Sperm Whale Myoglobin, Room Temperature Structure, Last 2 Degrees Of 5 Degree Total Oscillation And 160 Kgy Dose
Organism: Physeter catodon
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-03-27 Classification: OXYGEN TRANSPORT Ligands: HEM, CMO, SO4 |
 |
Organism: Candidatus pelagibacter ubique
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2011-12-28 Classification: TRANSFERASE Ligands: GOL, NA |
 |
Organism: Candidatus pelagibacter ubique
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2011-12-28 Classification: TRANSFERASE Ligands: DQY, GOL, NA |
 |
Organism: Candidatus pelagibacter ubique
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2011-12-28 Classification: TRANSFERASE Ligands: THG, ACT, GOL, NA |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2008-10-21 Classification: OXIDOREDUCTASE Ligands: FE2, 2CO |
 |
Organism: Caenorhabditis elegans
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2005-05-17 Classification: TRANSFERASE Ligands: GSH |

