Search Count: 73
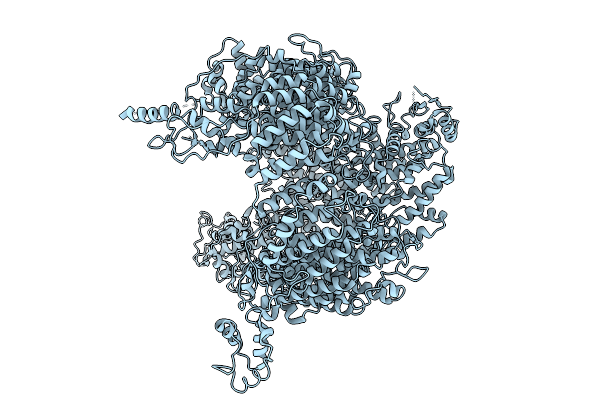 |
E3 Ubiquitin Ligase Huwe1 Homolog Tom1P In Closed-Conformation With Internal Acidic Domain Deletion
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: GENE REGULATION |
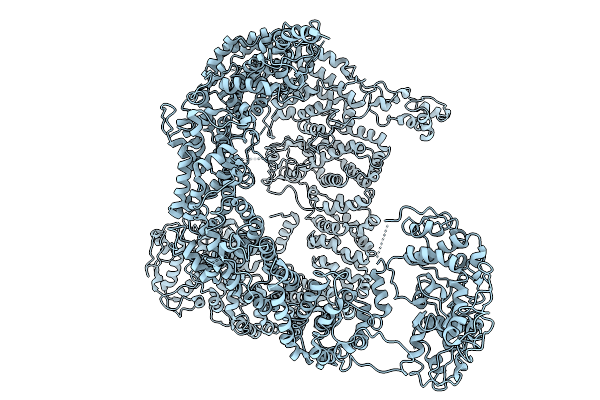 |
Full-Length And Internally His-Tagged Yeast E3 Ubiquitin Ligase Tom1P In An Open-Conformation
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: TRANSFERASE |
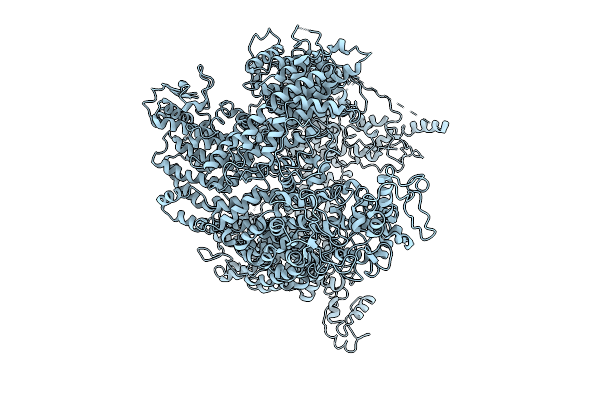 |
Cryo-Em Structure Of Full-Length And Internally His-Tagged Yeast E3 Ubiquitin Ligase Tom1P, In A Closed-Conformation State With Helical Repeat Solenoid Architecture
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: GENE REGULATION |
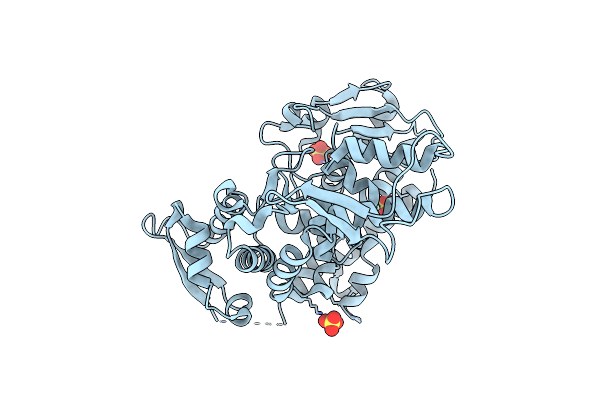 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Release Date: 2025-05-28 Classification: LIGASE Ligands: SO4 |
 |
Organism: Elysia chlorotica
Method: ELECTRON MICROSCOPY Release Date: 2025-01-29 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Elysia chlorotica
Method: ELECTRON MICROSCOPY Release Date: 2025-01-29 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Elysia chlorotica
Method: ELECTRON MICROSCOPY Resolution:3.03 Å Release Date: 2025-01-29 Classification: BIOSYNTHETIC PROTEIN Ligands: MLC |
 |
Organism: Elysia chlorotica
Method: ELECTRON MICROSCOPY Resolution:2.92 Å Release Date: 2025-01-29 Classification: BIOSYNTHETIC PROTEIN Ligands: NAP |
 |
Organism: Elysia chlorotica
Method: ELECTRON MICROSCOPY Resolution:3.15 Å Release Date: 2025-01-29 Classification: BIOSYNTHETIC PROTEIN Ligands: MLC, NAP |
 |
Organism: Elysia chlorotica
Method: ELECTRON MICROSCOPY Resolution:2.92 Å Release Date: 2025-01-29 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Elysia chlorotica
Method: ELECTRON MICROSCOPY Resolution:3.11 Å Release Date: 2025-01-29 Classification: BIOSYNTHETIC PROTEIN Ligands: NDP, 6VG |
 |
Organism: Elysia chlorotica
Method: ELECTRON MICROSCOPY Resolution:3.10 Å Release Date: 2025-01-29 Classification: BIOSYNTHETIC PROTEIN Ligands: NDP, 6VG |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: CHAPERONE Ligands: 08T, MG, ADP |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: CHAPERONE Ligands: 08T, MG, ADP |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: CHAPERONE Ligands: 08T, MG, ADP |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: CHAPERONE Ligands: 08T, MG, ADP |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: CHAPERONE Ligands: 08T, MG, ADP |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: CHAPERONE Ligands: 08T, MG, ADP |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: CHAPERONE Ligands: 08T, MG, ADP |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: CHAPERONE Ligands: 08T, MG, ADP |

