Search Count: 34
 |
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: VIRAL PROTEIN Ligands: A1CH4 |
 |
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: VIRAL PROTEIN Ligands: A1CH5 |
 |
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: VIRAL PROTEIN Ligands: A1CH6 |
 |
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: VIRAL PROTEIN Ligands: A1CH7 |
 |
Crystal Structure Of The Hiv Capsid Hexamer Bound To The Small Molecule Long-Acting Inhibitor, Gs-6207
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-07-01 Classification: VIRAL PROTEIN Ligands: QNG |
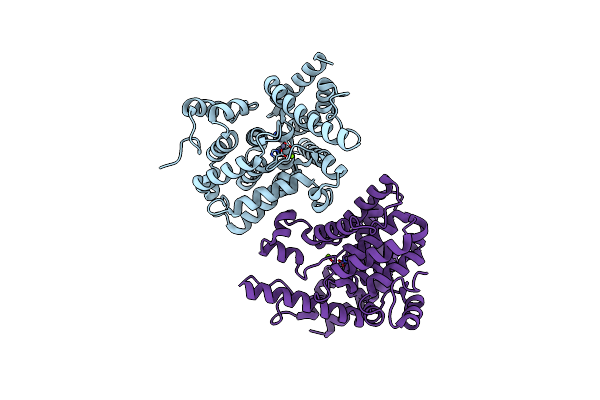 |
Organism: Schistosoma mansoni
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2019-01-30 Classification: HYDROLASE Ligands: ZN, MG |
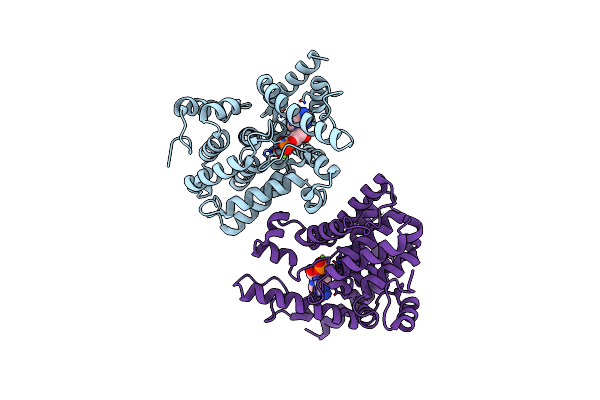 |
Organism: Schistosoma mansoni
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2018-12-12 Classification: HYDROLASE Ligands: ZN, MG, CMP, AMP |
 |
 |
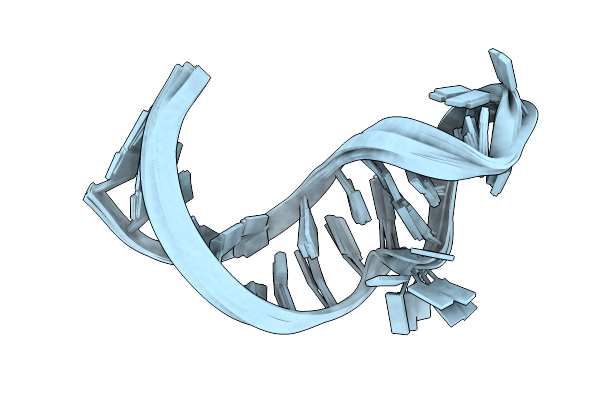
Crystal Structure Of Double-Stranded Rna With Four Terminal Gu Wobble Base Pairs
Organism: Endothia gyrosa
Method: X-RAY DIFFRACTION Resolution:1.32 Å Release Date: 2015-10-14 Classification: RNA Ligands: NCO, CL |
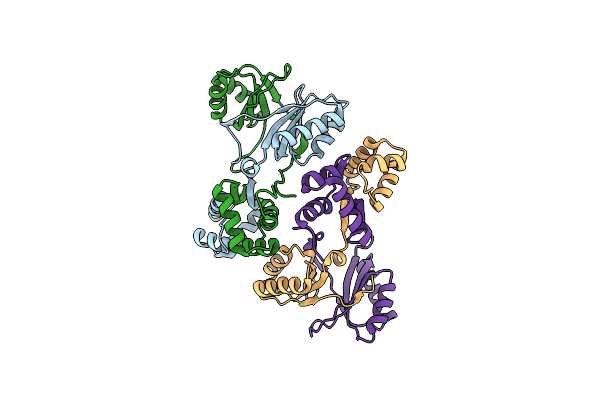 |
Sirohaem Decarboxylase Ahba/B - An Enzyme With Structural Homology To The Lrp/Asnc Transcription Factor Family That Is Part Of The Alternative Haem Biosynthesis Pathway.
Organism: Desulfovibrio desulfuricans
Method: X-RAY DIFFRACTION Resolution:2.23 Å Release Date: 2014-06-11 Classification: LYASE |
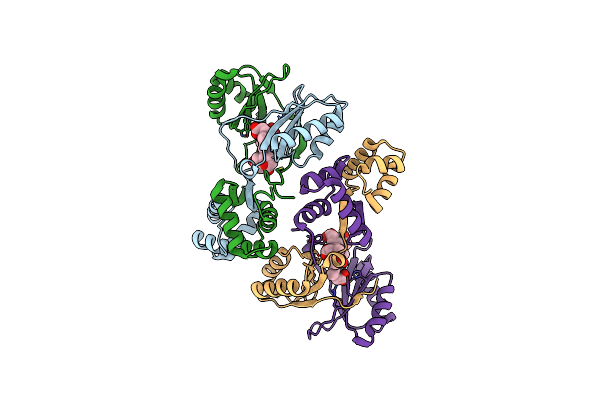 |
Sirohaem Decarboxylase Ahba/B - An Enzyme With Structural Homology To The Lrp/Asnc Transcription Factor Family That Is Part Of The Alternative Haem Biosynthesis Pathway.
Organism: Desulfovibrio desulfuricans
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2014-06-11 Classification: OXIDOREDUCTASE Ligands: OBV |
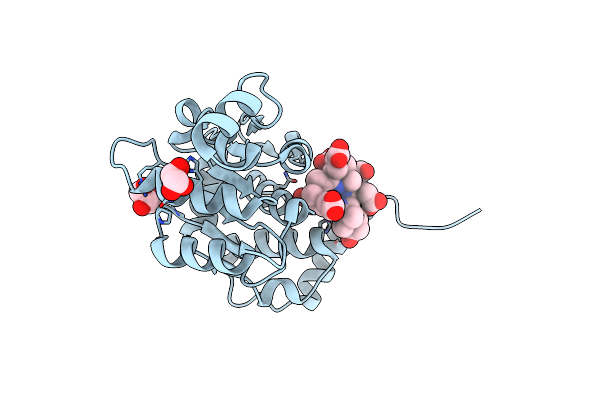 |
Organism: Rhodobacter capsulatus
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2012-10-17 Classification: ISOMERASE Ligands: 0UK, GOL |
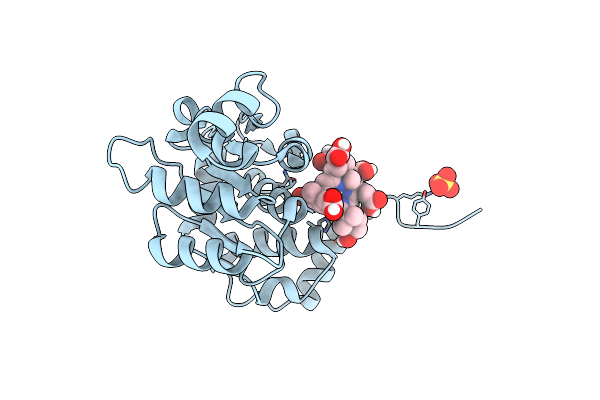 |
Crystal Structure Of Cobh (Precorrin-8X Methyl Mutase) Complexed With C5 Desmethyl-Hba
Organism: Rhodobacter capsulatus
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2012-09-19 Classification: ISOMERASE Ligands: P8X, SO4 |
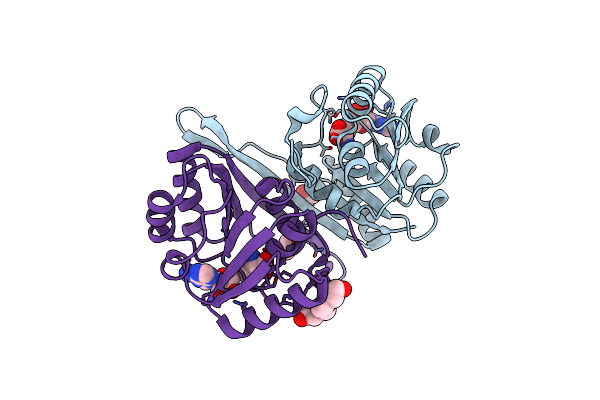 |
Crystal Structure Of C-Terminal Domain Of Precorrin-6Y C5,15-Methyltransferase From Rhodobacter Capsulatus
Organism: Rhodobacter capsulatus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2011-06-08 Classification: TRANSFERASE Ligands: SAH, GOL, PG4 |
 |
 |
Negamycin Binds To The Wall Of The Nascent Chain Exit Tunnel Of The 50S Ribosomal Subunit
Organism: Haloarcula marismortui
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2008-09-30 Classification: RIBOSOME Ligands: MG, K, NA, CL, NEG, CD |
 |
The Refined Crystal Structure Of The Haloarcula Marismortui Large Ribosomal Subunit At 2.4 Angstrom Resolution With Rrna Sequence For The 23S Rrna And Genome-Derived Sequences For R-Proteins
Organism: Haloarcula marismortui
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2008-05-20 Classification: RIBOSOME Ligands: MG, NA, CL, CD, K |
 |
Organism: Haloarcula marismortui
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2008-05-20 Classification: RIBOSOME Ligands: MG, CL, SR, NA, CD, K, ANM |
 |
Structure Of Anisomycin Resistant 50S Ribosomal Subunit: 23S Rrna Mutation C2487U
Organism: Haloarcula marismortui
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2008-05-20 Classification: RIBOSOME Ligands: MG, CL, SR, NA, CD, K |
 |
Structure Of Anisomycin Resistant 50S Ribosomal Subunit: 23S Rrna Mutation U2535A
Organism: Haloarcula marismortui
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2008-05-20 Classification: RIBOSOME Ligands: MG, CL, SR, NA, CD, K |

