Search Count: 25
 |
Organism: Homo sapiens, Influenza a virus (a/zhejiang/dtid-zju01/2013(h7n9)), Synthetic construct, Influenza b virus, Sus scrofa domesticus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: VIRUS Ligands: ZN, G1G, MG, PO4 |
 |
Organism: Homo sapiens, Influenza a virus (a/zhejiang/dtid-zju01/2013(h7n9)), Influenza b virus, Synthetic construct, Sus scrofa domesticus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: VIRUS Ligands: ZN, MG, G1G, PO4 |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: VIRAL PROTEIN Ligands: 6CJ |
 |
Organism: Saccharomyces cerevisiae, Human immunodeficiency virus 2
Method: ELECTRON MICROSCOPY Release Date: 2024-10-30 Classification: GENE REGULATION Ligands: ZN, MG |
 |
Organism: Saccharomyces cerevisiae, Human immunodeficiency virus 2
Method: ELECTRON MICROSCOPY Release Date: 2024-10-30 Classification: GENE REGULATION |
 |
Organism: Saccharomyces cerevisiae, Human immunodeficiency virus 2
Method: ELECTRON MICROSCOPY Release Date: 2024-10-30 Classification: GENE REGULATION Ligands: ADP, MG, BEF |
 |
Organism: Saccharomyces cerevisiae, Human immunodeficiency virus 2
Method: ELECTRON MICROSCOPY Release Date: 2024-10-30 Classification: GENE REGULATION Ligands: ZN, MG, ADP, BEF |
 |
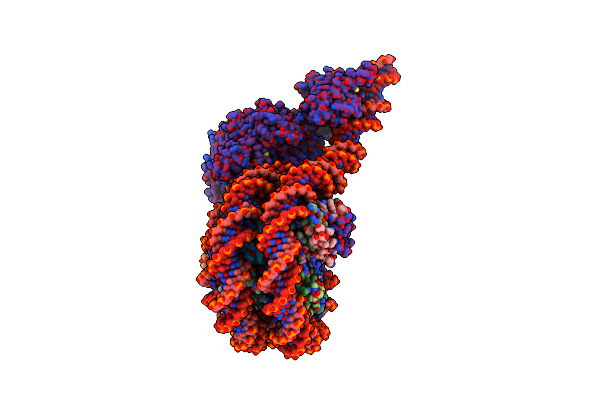
Structure Of A Hexasome-Nucleosome Complex With A Dyad-To-Dyad Distance Of 103 Bp.
Organism: Xenopus laevis, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-09-18 Classification: DNA BINDING PROTEIN |
 |
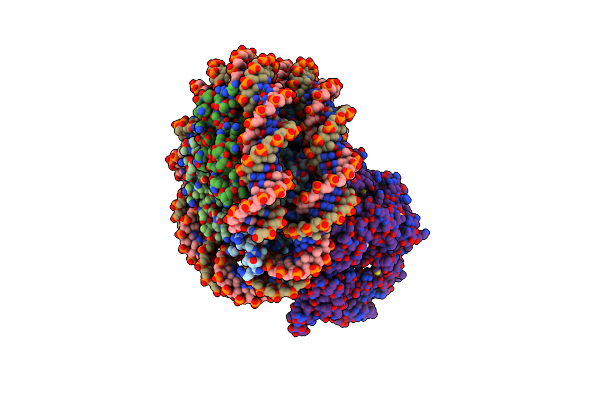
Structure Of Chd1 Bound To A Hexasome-Nucleosome Complex With A Dyad-To-Dyad Distance Of 103 Bp.
Organism: Xenopus laevis, Synthetic construct, Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2024-09-18 Classification: DNA BINDING PROTEIN Ligands: ADP, BEF, MG |
 |
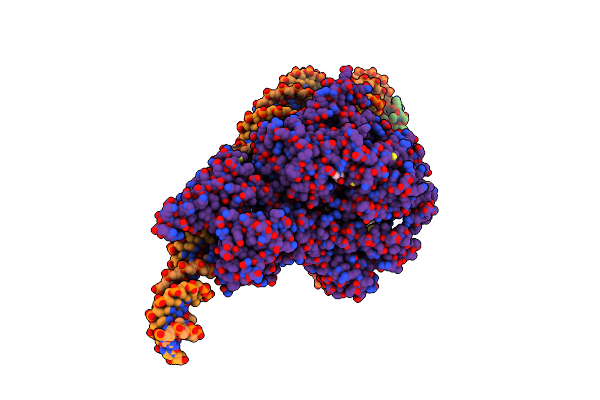
Structure Of Chd1 Bound To A Dinucleosome With A Dyad-To-Dyad Distance Of 103 Bp.
Organism: Xenopus laevis, Synthetic construct, Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2024-09-18 Classification: DNA BINDING PROTEIN Ligands: ADP, BEF, MG |
 |
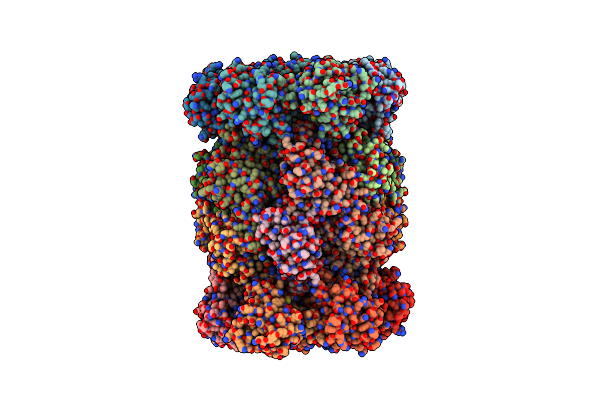
Structure Of A Mononucleosome Bound By One Copy Of Chd1 With The Dbd On The Exit-Side Dna.
Organism: Xenopus laevis, Synthetic construct, Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2024-09-18 Classification: DNA BINDING PROTEIN Ligands: ADP, BEF, MG |
 |
Yeast 20S Proteasome In Complex With An Engineered Fellutamide Derivative (C14Qal)
Organism: Synthetic construct, Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.25 Å Release Date: 2023-12-20 Classification: HYDROLASE Ligands: MG, CL, MYR |
 |
Rna Polymerase Ii Pre-Initiation Complex With The Distal +1 Nucleosome (Pic-Nuc18W)
Organism: Homo sapiens, Xenopus laevis, Unidentified adenovirus, Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2023-05-03 Classification: TRANSCRIPTION Ligands: SF4, ZN, MG |
 |
Rna Polymerase Ii Pre-Initiation Complex With The Proximal +1 Nucleosome (Pic-Nuc10W)
Organism: Homo sapiens, Xenopus laevis, Unidentified adenovirus, Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2023-05-03 Classification: TRANSCRIPTION Ligands: SF4, ZN, MG |
 |
Rna Polymerase Ii Core Pre-Initiation Complex With The Proximal +1 Nucleosome (Cpic-Nuc10W)
Organism: Homo sapiens, Xenopus laevis, Unidentified adenovirus, Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2023-05-03 Classification: TRANSCRIPTION Ligands: ZN, MG |
 |
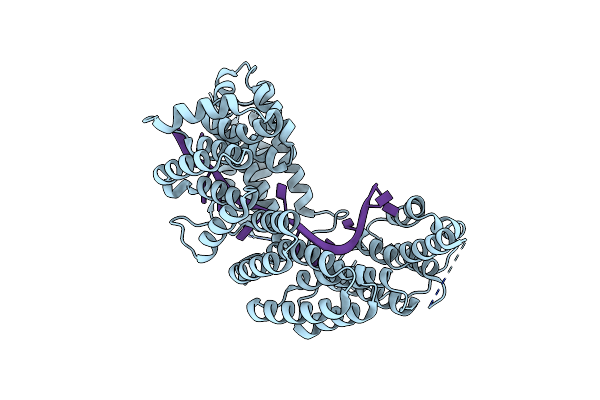
Organism: Saccharomyces cerevisiae s288c, Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2021-04-07 Classification: RNA BINDING PROTEIN Ligands: CL |
 |
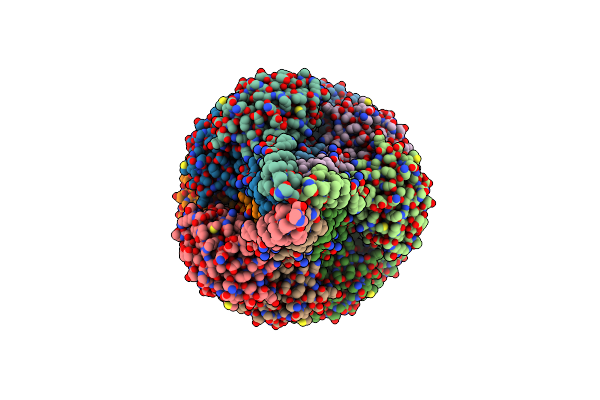
Organism: Saccharomyces cerevisiae s288c, Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2021-04-07 Classification: RNA BINDING PROTEIN Ligands: CL |
 |
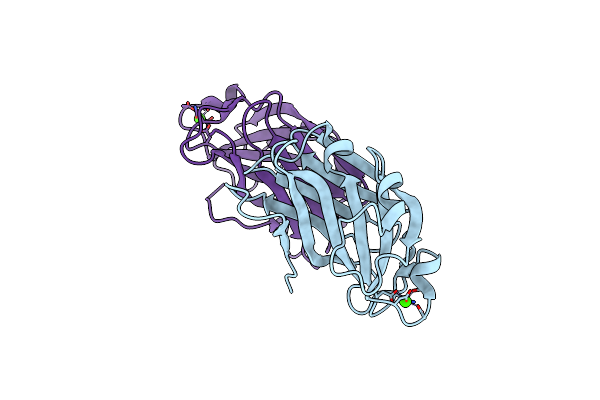
Organism: Methanococcus maripaludis (strain s2 / ll)
Method: ELECTRON MICROSCOPY Release Date: 2019-02-13 Classification: PROTEIN FIBRIL |
 |
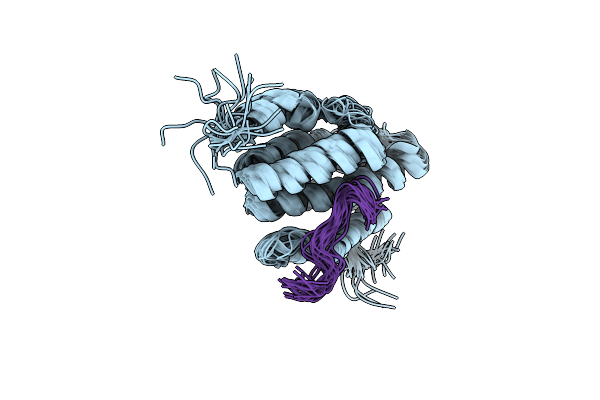
Crystal Structure Of Archaeal Flagellin Flab1 From Methanocaldococcus Jannaschii
Organism: Methanocaldococcus jannaschii dsm 2661
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2019-02-06 Classification: STRUCTURAL PROTEIN Ligands: CA |
 |
Organism: Saccharomyces cerevisiae
Method: SOLUTION NMR Release Date: 2014-08-13 Classification: TRANSCRIPTION |

