Search Count: 156
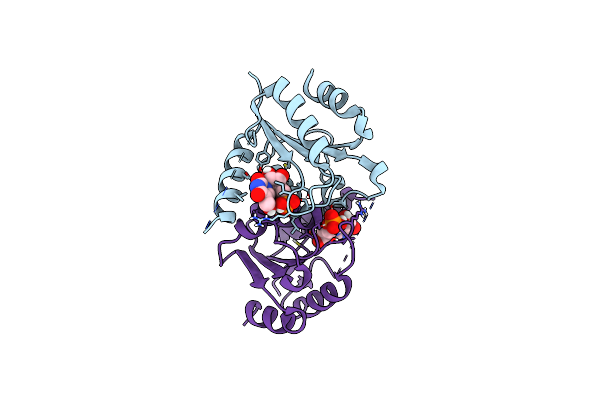 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2025-02-05 Classification: HYDROLASE/INHIBITOR Ligands: A1BBB |
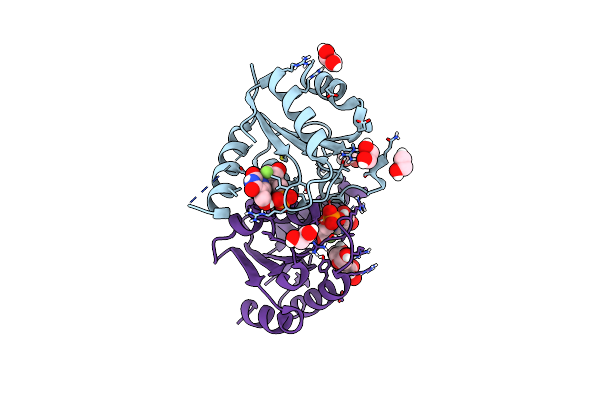 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.13 Å Release Date: 2025-02-05 Classification: HYDROLASE/INHIBITOR Ligands: A1BBC, EDO, PEG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2025-02-05 Classification: HYDROLASE/INHIBITOR Ligands: A1BBD |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2025-02-05 Classification: HYDROLASE/INHIBITOR Ligands: A1BBE |
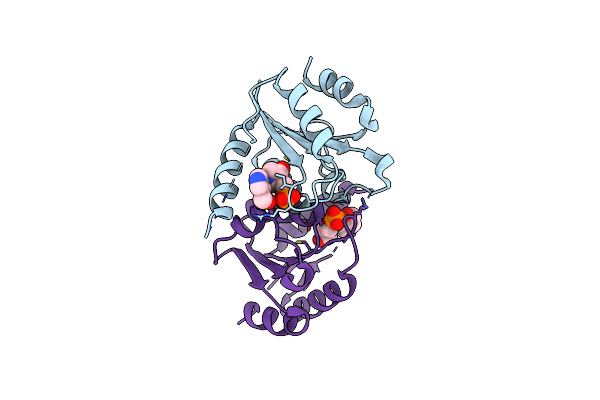 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.82 Å Release Date: 2025-02-05 Classification: HYDROLASE/INHIBITOR Ligands: A1BBF |
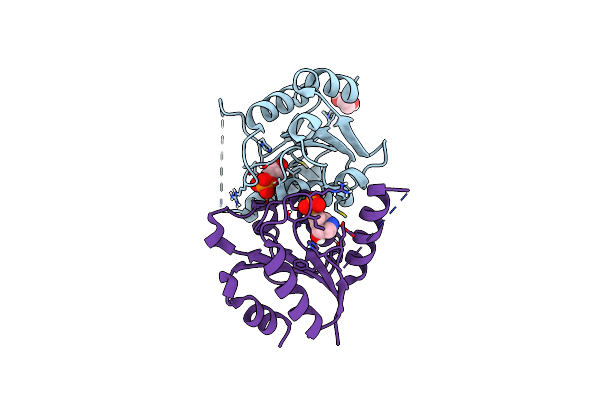 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2025-02-05 Classification: HYDROLASE/INHIBITOR Ligands: NR1, EDO |
 |
Organism: Kluyveromyces lactis nrrl y-1140
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-10-18 Classification: TRANSFERASE Ligands: HPA, ACT |
 |
Structure Of K. Lactis Pnp Bound To Transition State Analog Dadme-Immucillin H And Sulfate
Organism: Kluyveromyces lactis nrrl y-1140
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2023-10-18 Classification: TRANSFERASE Ligands: DIH, SO4, GOL |
 |
Structure Of K. Lactis Pnp S42E Variant Bound To Transition State Analog Dadme-Immucillin G And Sulfate
Organism: Kluyveromyces lactis nrrl y-1140
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-10-18 Classification: TRANSFERASE Ligands: IM5, SO4, GUN |
 |
Structure Of K. Lactis Pnp S42E-H98R Variant Bound To Transition State Analog Dadme-Immucillin G And Sulfate
Organism: Kluyveromyces lactis nrrl y-1140
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2023-10-18 Classification: TRANSFERASE Ligands: IM5, SO4 |
 |
Structure Of Bacteroides Fragilis Pnp Bound To Transition State Analog Immucillin H And Sulfate
Organism: Bacteroides fragilis nctc 9343
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2023-10-18 Classification: TRANSFERASE Ligands: IMH, SO4 |
 |
Structure Of Clostridium Perfringens Pnp Bound To Transition State Analog Immucillin H And Sulfate
Organism: Clostridium perfringens atcc 13124
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2023-10-18 Classification: TRANSFERASE Ligands: IMH, SO4 |
 |
Crystal Structure Of Human Phenylethanolamine N-Methyltransferase (Pnmt) In Complex With (2S)-2-Amino-4-(((5-(6-Amino-9H-Purin-9-Yl)-3,4-Dihydroxytetrahydrofuran-2-Yl)Methyl)(4-(7,8-Dichloro-1,2,3,4-Tetrahydroisoquinolin-4-Yl)Butyl)Amino)Butanoic Acid And Adohcy (Sah)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-02-15 Classification: TRANSFERASE Ligands: SAH, CD, EDO, KNX |
 |
Crystal Structure Of Human Phenylethanolamine N-Methyltransferase (Pnmt) In Complex With (2S)-2-Amino-4-(((5-(6-Amino-9H-Purin-9-Yl)-3,4-Dihydroxytetrahydrofuran-2-Yl)Methyl)(4-(7,8-Dichloro-1,2,3,4-Tetrahydroisoquinolin-4-Yl)Butyl)Amino)Butanoic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2023-02-15 Classification: TRANSFERASE Ligands: EDO, KNX |
 |
Crystal Structure Of Plasmodium Falciparum Hypoxanthine-Guanine-Xanthine Phosphoribosyltransferase In Complex With [(3S)-4-Hydroxy-3-[({2-Amino-4-Hydroxy-5H-Pyrrolo[3,2-D]Pyrimidin-7-Yl}Methyl)Amino]Butyl]Phosphonic Acid
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2022-12-07 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: YBM, POP, MG, EDO, ACY |
 |
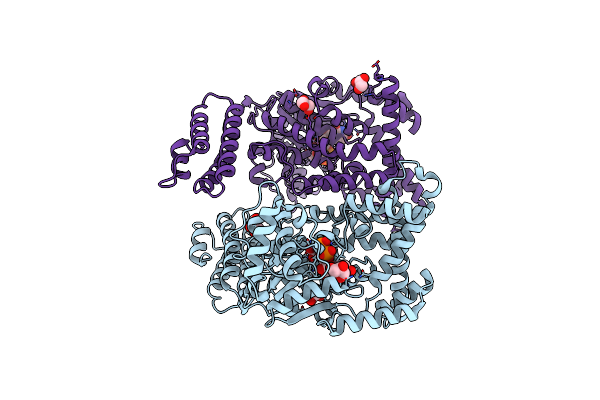
Crystal Structure Of Clostridium Difficile Toxin B (Tcdb) Glucosyltransferase In Complex With Udp And Isofagomine
Organism: Clostridioides difficile
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2021-11-17 Classification: TRANSFERASE/Inhibitor Ligands: EDO, UDP, IFM, MN |
 |
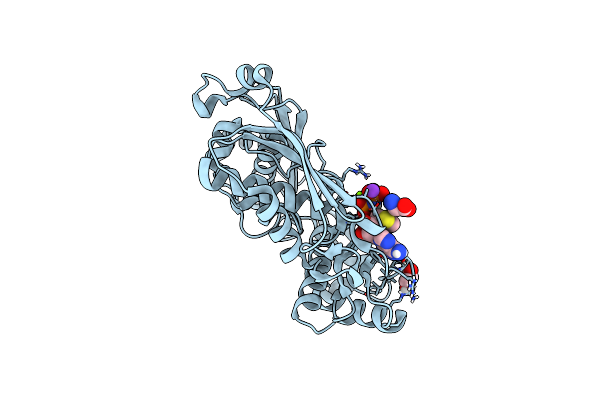
Crystal Structure Of Clostridium Difficile Toxin B (Tcdb) Glucosyltransferase In Complex With Udp And Noeuromycin
Organism: Clostridioides difficile
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-11-17 Classification: TRANSFERASE/Inhibitor Ligands: NOY, UDP, GOL, MN |
 |
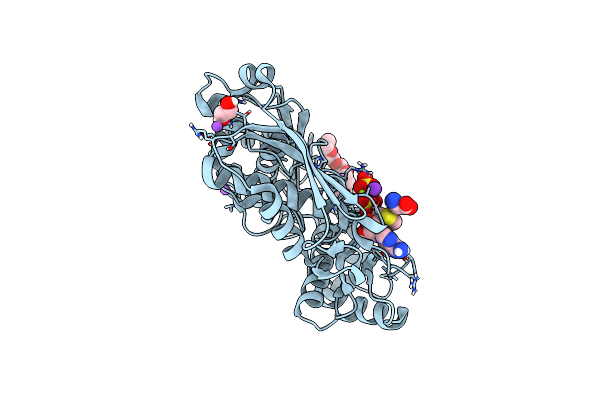
Human Methionine Adenosyltransferase 2A Bound To Methylthioadenosine And Inhibitor Imido-Diphosphate (Pnp)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.07 Å Release Date: 2021-11-10 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: MTA, ALA, 2PN, EDO, MG, K |
 |
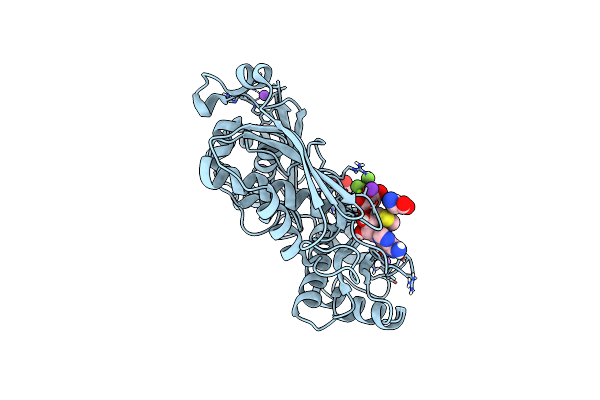
Human Methionine Adenosyltransferase 2A Bound To Methylthioadenosine And Two Sulfate In The Active Site
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2021-11-10 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: ALA, MTA, P3G, EDO, SO4, MG, K, NA |
 |
Human Methionine Adenosyltransferase 2A Bound To Methylthioadenosine, Malonate (Mla) And Mgf3
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.07 Å Release Date: 2021-11-03 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: MTA, ALA, MLA, MGF, GOL, MG, K, NA |

