Search Count: 28
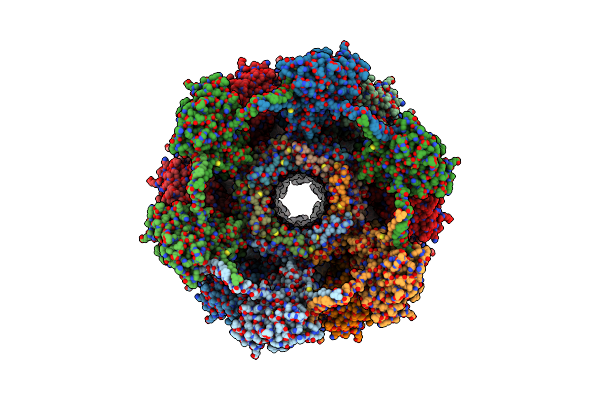 |
Organism: Clostridioides difficile
Method: ELECTRON MICROSCOPY Release Date: 2024-08-28 Classification: VIRUS LIKE PARTICLE |
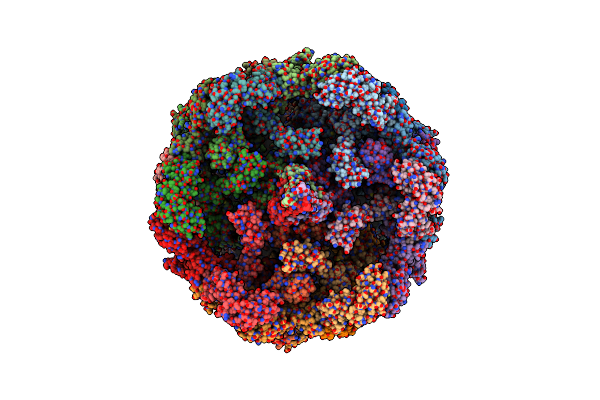 |
Cryoem Structure Of Diffocin - Precontracted - Baseplate - Focused Refinement On Triplex Region
Organism: Clostridioides difficile
Method: ELECTRON MICROSCOPY Release Date: 2024-08-28 Classification: VIRUS LIKE PARTICLE |
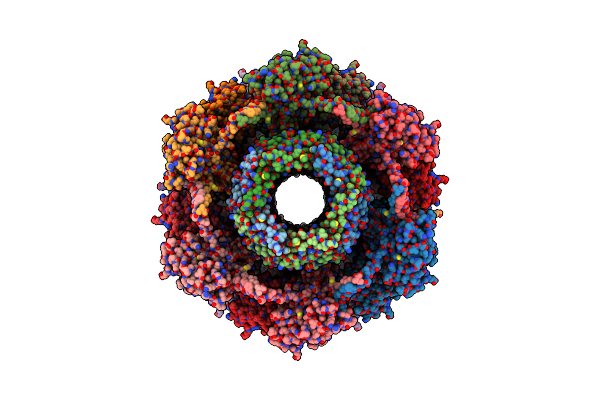 |
Organism: Clostridioides difficile
Method: ELECTRON MICROSCOPY Release Date: 2024-08-28 Classification: VIRUS LIKE PARTICLE |
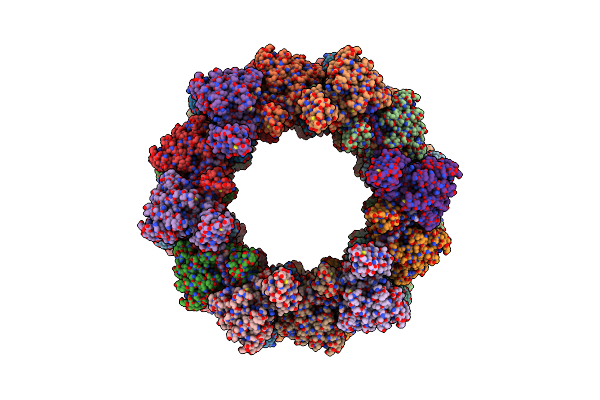 |
Organism: Clostridioides difficile
Method: ELECTRON MICROSCOPY Release Date: 2024-08-28 Classification: VIRUS LIKE PARTICLE |
 |
Cryoem Structure Of Diffocin - Postcontracted - Collar - Transitional State
Organism: Clostridioides difficile
Method: ELECTRON MICROSCOPY Release Date: 2024-08-28 Classification: VIRUS LIKE PARTICLE |
 |
Organism: Clostridioides difficile
Method: ELECTRON MICROSCOPY Release Date: 2024-08-28 Classification: VIRUS LIKE PARTICLE |
 |
Cryoem Structure Of Diffocin - Postcontracted - Baseplate - Transitional State
Organism: Clostridioides difficile
Method: ELECTRON MICROSCOPY Release Date: 2024-08-28 Classification: VIRUS LIKE PARTICLE |
 |
Organism: Clostridioides difficile
Method: ELECTRON MICROSCOPY Release Date: 2024-08-28 Classification: VIRUS LIKE PARTICLE |
 |
Organism: Homo sapiens, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.71 Å Release Date: 2021-06-30 Classification: HYDROLASE Ligands: ATP, EDO, GOL |
 |
Structure Of The N-Terminal Catalytic Region Of T. Thermophilus Rel Bound To Ppgpp
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2020-07-08 Classification: TRANSFERASE Ligands: G4P, GOL, MN, MG, CL, NA |
 |
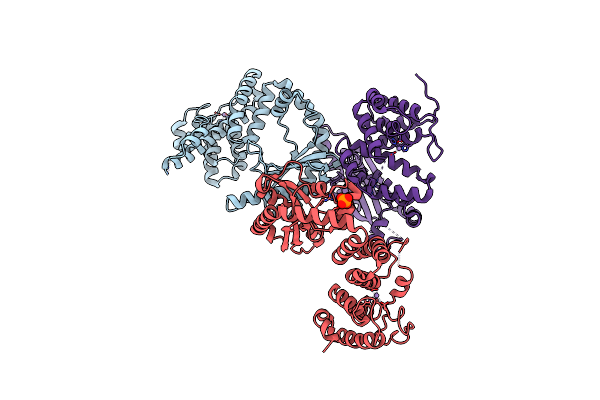
Structure Of The Catalytic Domain Of T. Thermophilus Rel In Complex With Amp And Ppgpp
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2020-07-08 Classification: TRANSFERASE Ligands: MN, AMP, GN3, TMO, MG |
 |
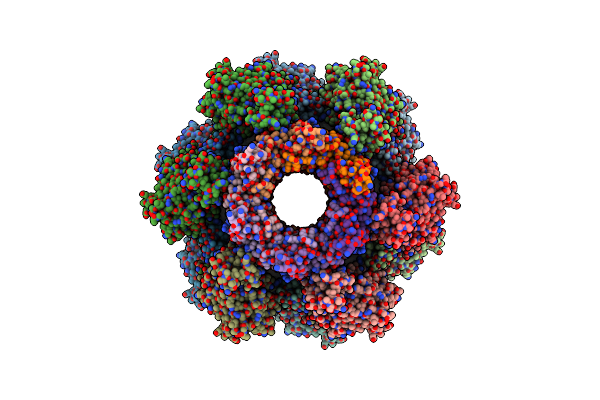
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.96 Å Release Date: 2020-07-08 Classification: TRANSFERASE Ligands: MN, CL, PO4, NA |
 |
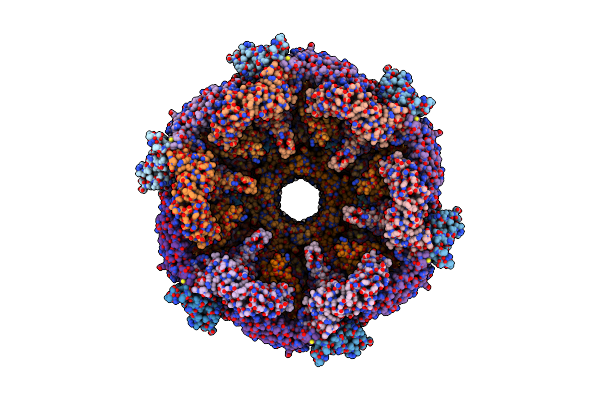
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: ELECTRON MICROSCOPY Release Date: 2020-04-15 Classification: ANTIMICROBIAL PROTEIN |
 |
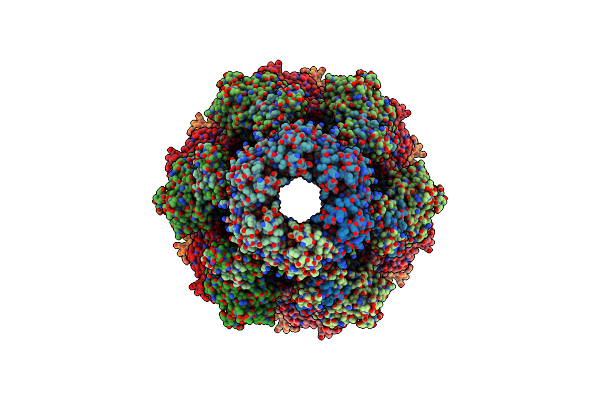
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: ELECTRON MICROSCOPY Release Date: 2020-04-15 Classification: ANTIMICROBIAL PROTEIN |
 |
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: ELECTRON MICROSCOPY Release Date: 2020-04-15 Classification: ANTIMICROBIAL PROTEIN |
 |
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: ELECTRON MICROSCOPY Release Date: 2020-04-15 Classification: UNKNOWN FUNCTION |
 |
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: ELECTRON MICROSCOPY Release Date: 2020-04-15 Classification: ANTIMICROBIAL PROTEIN |
 |
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: ELECTRON MICROSCOPY Release Date: 2020-04-15 Classification: ANTIMICROBIAL PROTEIN |
 |
Organism: Homo sapiens, Lama glama
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2019-06-26 Classification: IMMUNE SYSTEM Ligands: MG, ATP |
 |
Organism: Homo sapiens, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2019-06-26 Classification: HYDROLASE Ligands: GOL, PEG |

