Search Count: 35
 |
Cryo-Em Structure Of Human Fibroblast Activation Protein Alpha Dimer With One Sumo-I3 Vhhs Bound
Organism: Homo sapiens, Lama glama
Method: ELECTRON MICROSCOPY Resolution:2.70 Å Release Date: 2025-02-26 Classification: HYDROLASE Ligands: NAG |
 |
Cryo-Em Structure Of Human Fibroblast Activation Protein Alpha Dimer With Two Sumo-I3 Vhhs Bound
Organism: Homo sapiens, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2025-02-26 Classification: HYDROLASE Ligands: NAG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-02-12 Classification: TRANSFERASE Ligands: MN, UGA |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-10-16 Classification: SIGNALING PROTEIN |
 |
Crystal Structure Of Pa0012 Complexed With Cyclic-Di-Gmp From Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa pa14
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2024-09-04 Classification: SIGNALING PROTEIN Ligands: C2E, GOL |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-08-14 Classification: RECOMBINATION |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-08-14 Classification: SIGNALING PROTEIN |
 |
Organism: Cryptococcus neoformans var. grubii h99
Method: ELECTRON MICROSCOPY Release Date: 2024-07-03 Classification: LIGASE |
 |
Cryoem Structure Of Cryptococcus Neoformans H99 Acetyl-Coa Synthetase In Complex With Adenosine-5'-Ethylphosphate
Organism: Cryptococcus neoformans var. grubii h99
Method: ELECTRON MICROSCOPY Release Date: 2024-07-03 Classification: LIGASE Ligands: WTA |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-02-14 Classification: PROTEIN TRANSPORT |
 |
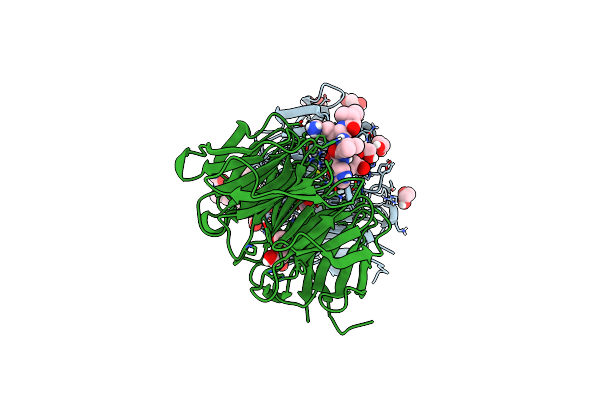
Crystal Structure Of Beta'-Copi-Wd40 Domain In Complex With Sars-Cov-2 Spike Tail Hepta-Peptide
Organism: Saccharomyces cerevisiae, Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2024-01-31 Classification: PROTEIN TRANSPORT |
 |
Crystal Structure Of Beta'-Copi-Wd40 Domain In Complex With Sars-Cov-2 Clientized Spike Tail Heptapeptide.
Organism: Saccharomyces cerevisiae, Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2024-01-31 Classification: PROTEIN TRANSPORT Ligands: EDO |
 |
Crystal Structure Of Beta'-Copi-Wd40 Domain Y33A Mutant In Complex With Sars-Cov-2 Clientized Spike Tail Heptapeptide.
Organism: Saccharomyces cerevisiae, Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-01-31 Classification: PROTEIN TRANSPORT |
 |
Organism: Schizosaccharomyces pombe
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-01-31 Classification: PROTEIN TRANSPORT Ligands: ACE |
 |
Organism: Schizosaccharomyces pombe
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2024-01-31 Classification: PROTEIN TRANSPORT Ligands: ACE |
 |
Organism: Schizosaccharomyces pombe
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-01-31 Classification: PROTEIN TRANSPORT Ligands: ACE |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-09-07 Classification: DNA BINDING PROTEIN/DNA |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-09-07 Classification: DNA BINDING PROTEIN/DNA |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-09-07 Classification: DNA BINDING PROTEIN/DNA |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-09-07 Classification: DNA BINDING PROTEIN/DNA |

