Search Count: 251
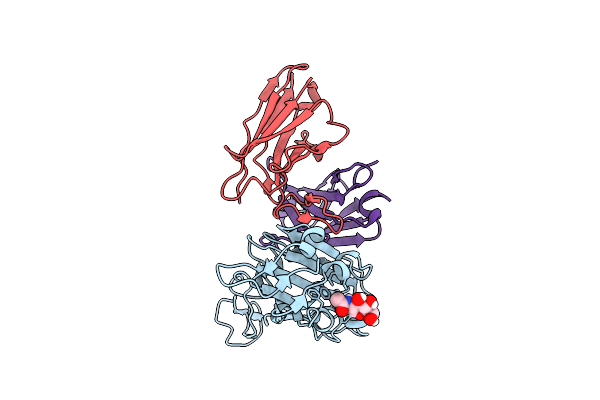 |
Organism: Severe acute respiratory syndrome coronavirus 2, Tequatrovirus t4, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-23 Classification: PROTEIN BINDING Ligands: NAG |
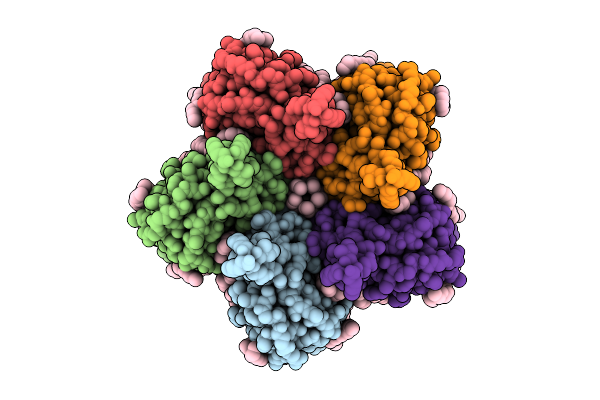 |
Cryo-Em Structure Of The Flotillin-Associated Rhodopsin Psfar In Detergent Micelle
Organism: Candidatus pseudothioglobus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-23 Classification: MEMBRANE PROTEIN Ligands: LFA |
 |
Cryo-Em Structure Of The Light-Driven Proton Pump Pspr In Detergent Micelle
Organism: Candidatus pseudothioglobus sp.
Method: ELECTRON MICROSCOPY Release Date: 2025-07-23 Classification: MEMBRANE PROTEIN Ligands: LFA, RET, LMT |
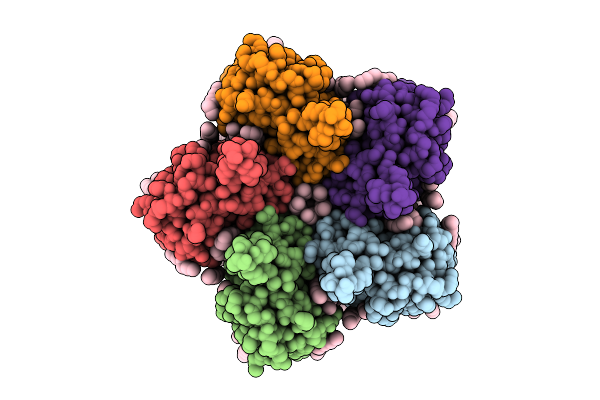 |
Cryo-Em Structure Of The Double Mutant H84V/E120G Of The Flotillin-Associated Rhodopsin Psfar In Detergent Micelle
Organism: Candidatus pseudothioglobus sp.
Method: ELECTRON MICROSCOPY Release Date: 2025-07-23 Classification: MEMBRANE PROTEIN Ligands: LFA, RET |
 |
Organism: Cryobacterium levicorallinum
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: MEMBRANE PROTEIN Ligands: LFA, RET |
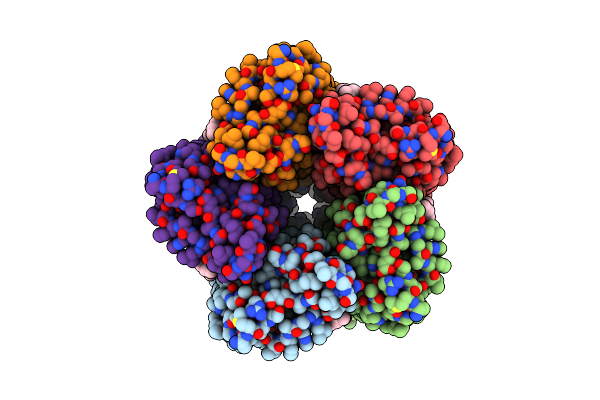 |
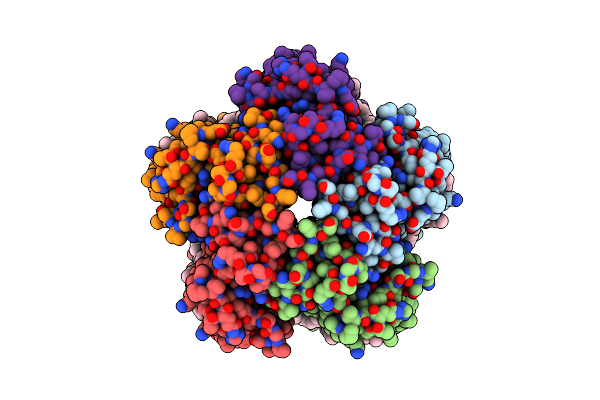
Organism: Cryobacterium levicorallinum
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: MEMBRANE PROTEIN Ligands: LFA, RET |
 |
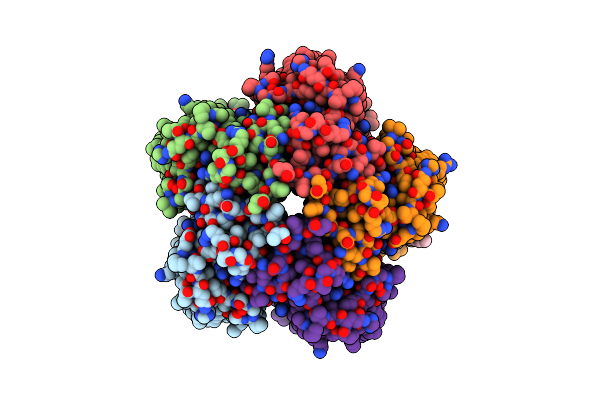
Organism: Cryobacterium levicorallinum
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: MEMBRANE PROTEIN Ligands: LMT, LFA, RET |
 |
Cryo-Em Structure Of The Microbial Rhodopsin Cryor1 At Ph 10.5 In Detergent In The Ground State
Organism: Cryobacterium levicorallinum
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: MEMBRANE PROTEIN Ligands: LMT, RET, LFA |
 |
Cryo-Em Structure Of The Microbial Rhodopsin Cryor1 At Ph 10.5 In Detergent In The M State
Organism: Cryobacterium levicorallinum
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: MEMBRANE PROTEIN Ligands: LMT, RET |
 |
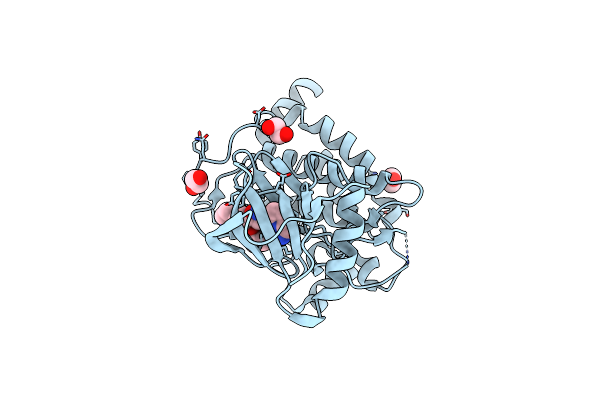
Organism: Subtercola endophyticus
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: MEMBRANE PROTEIN Ligands: LFA, RET |
 |
Crystal Structure Of Human Tak1/Tab1 Fusion Protein In Complex With Compound S1
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2024-12-04 Classification: TRANSFERASE Ligands: A1IED, EDO |
 |
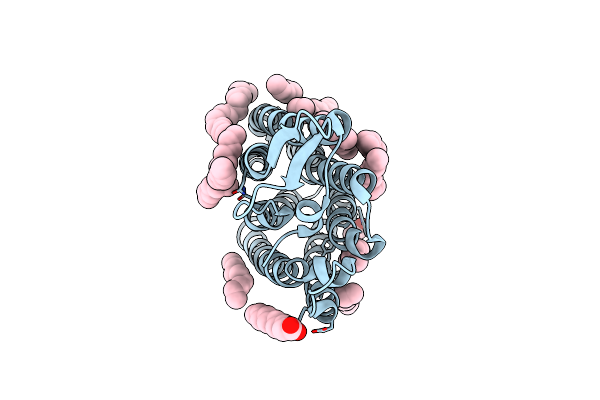
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.11 Å Release Date: 2024-10-30 Classification: PROTEIN BINDING |
 |
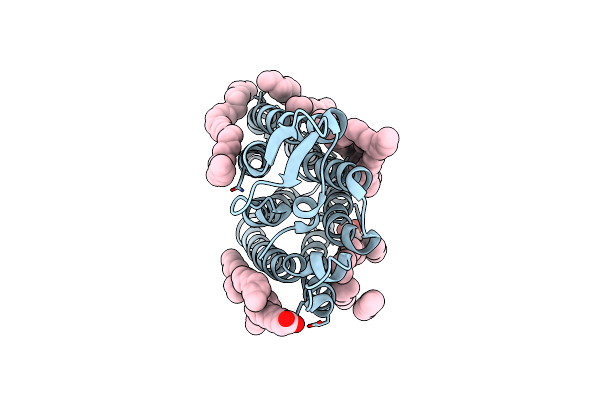
Crystal Structure Of The Light-Driven Sodium Pump Ernar In The Monomeric Form At Ph 4.6
Organism: Erythrobacter
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-04-24 Classification: MEMBRANE PROTEIN Ligands: LFA, OLA |
 |
Crystal Structure Of The Light-Driven Sodium Pump Ernar In The Monomeric Form At Ph 8.8
Organism: Erythrobacter
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2024-04-24 Classification: MEMBRANE PROTEIN Ligands: LFA, OLA |
 |
Cryo-Em Structure Of The Light-Driven Sodium Pump Ernar In The Pentameric Form At Ph 8.0
Organism: Erythrobacter
Method: ELECTRON MICROSCOPY Resolution:2.63 Å Release Date: 2024-04-24 Classification: MEMBRANE PROTEIN Ligands: LFA, LMT |
 |
Cryo-Em Structure Of The Light-Driven Sodium Pump Ernar In The Pentameric Form At Ph 4.3
Organism: Erythrobacter
Method: ELECTRON MICROSCOPY Resolution:2.50 Å Release Date: 2024-04-24 Classification: MEMBRANE PROTEIN Ligands: LMT, LFA |
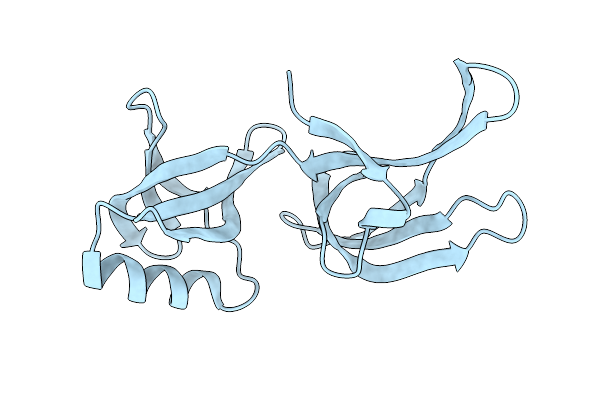 |
Structure Of Hex-1 From N. Crassa Crystallized In Cellulo (Cytosol), Diffracted At 100K And Resolved Using Crystfel
Organism: Neurospora crassa
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2024-02-21 Classification: STRUCTURAL PROTEIN |
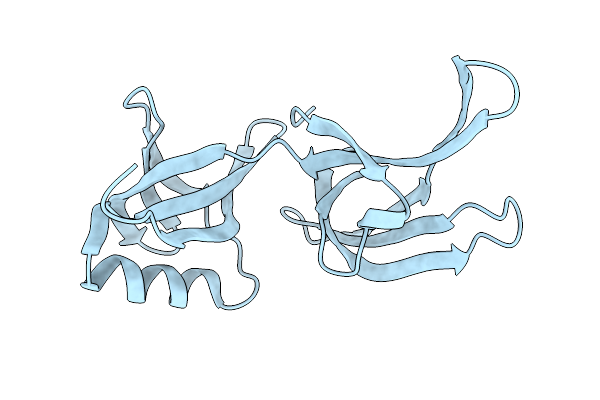 |
Structure Of Hex-1 From N. Crassa Crystallized In Cellulo, Diffracted At 100K And Resolved Using Crystfel
Organism: Neurospora crassa
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2024-02-21 Classification: STRUCTURAL PROTEIN |
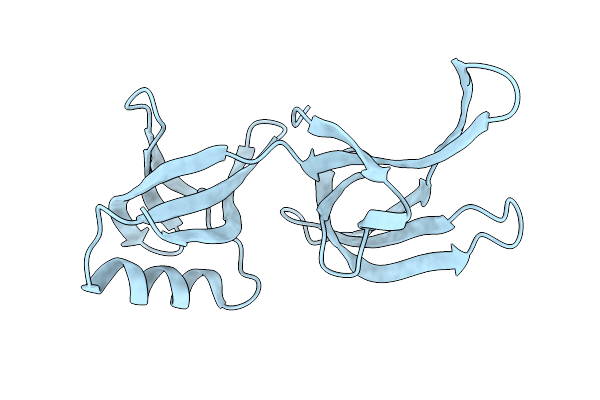 |
Structure Of Hex-1 (Cyto V2) From N. Crassa Grown In Living Insect Cells, Diffracted At 100K And Resolved Using Crystfel
Organism: Neurospora crassa
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-02-21 Classification: STRUCTURAL PROTEIN |
 |
Structure Of Hex-1 From N. Crassa Crystallized In Cellulo, Diffracted At 100K And Resolved Using Xds
Organism: Neurospora crassa
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-02-21 Classification: STRUCTURAL PROTEIN |

