Search Count: 38
 |
Crystal Structure Of The Putative Oxidoreductase Of Duf1479-Containing Protein Family Ypo2976 From Yersinia Pestis Bound To Caps
Organism: Yersinia pestis co92
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2022-02-16 Classification: UNKNOWN FUNCTION Ligands: CXS, MG, PO4, SO4, GOL |
 |
Crystal Structure Of The Putative Oxidoreductase Of Duf1479-Containing Protein Family Ypo2976 From Yersinia Pestis Bound To 2-Oxo-Glutaric Acid
Organism: Yersinia pestis co92
Method: X-RAY DIFFRACTION Resolution:2.41 Å Release Date: 2022-02-16 Classification: UNKNOWN FUNCTION Ligands: MN, AKG, EDO |
 |
Crystal Structure Of The Dna-Binding Domain Of The Lysr Family Transcriptional Regulator Yfba From Yersinia Pestis
Organism: Yersinia pestis co92
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-02-09 Classification: TRANSCRIPTION Ligands: SO4, CL, GOL, FMT |
 |
Crystal Structure Of The C-Terminal Ligand-Binding Domain Of The Lysr Family Transcriptional Regulator Yfba From Yersinia Pestis
Organism: Yersinia pestis co92
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2022-02-09 Classification: TRANSCRIPTION Ligands: 3HB, PO4 |
 |
Crystal Structure Of The Protein Of Unknown Function Ypo0625 From Yersinia Pestis
Organism: Yersinia pestis co92
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2022-02-02 Classification: UNKNOWN FUNCTION Ligands: PEG, K, CL, EDO, GOL |
 |
Crystal Structure Of The Soluble Domain Of The Putative Ompa -Family Membrane Protein Ypo0514 From Yersinia Pestis
Organism: Yersinia pestis co92
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-01-26 Classification: TRANSPORT PROTEIN Ligands: CA, PO4 |
 |
Crystal Structure Of The Putative Exported Protein Ypo2471 From Yersinia Pestis
Organism: Yersinia pestis co92
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2022-01-19 Classification: UNKNOWN FUNCTION Ligands: CL, EDO, ACY, GOL |
 |
Crystal Structure Of The Pirin Family Protein Redox-Sensitive Bicupin Yhak Bound To Copper Ion From Yersinia Pestis
Organism: Yersinia pestis co92
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2022-01-19 Classification: METAL BINDING PROTEIN Ligands: CU, EDO, SIN, FMT |
 |
Crystal Structure Of The Pirin Family Protein Redox-Sensitive Bicupin Yhak In The Presence Of Fe Ion From Yersinia Pestis
Organism: Yersinia pestis co92
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2022-01-19 Classification: METAL BINDING PROTEIN Ligands: FE, CL |
 |
Crystal Structure Of The Pirin Family Protein Redox-Sensitive Bicupin Yhak From Yersinia Pestis
Organism: Yersinia pestis co92
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2022-01-12 Classification: METAL BINDING PROTEIN Ligands: MG |
 |
Crystal Structure Of The Protein Of Unknown Function Of The Conserved Rid Protein Family Yyfa From Yersinia Pestis
Organism: Yersinia pestis
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2017-04-05 Classification: Structural genomics, unknown function Ligands: ACY, CA, GOL |
 |
Crystal Structure Of The Protein Of Unknown Function Of The Conserved Rid Protein Family Yyfb From Yersinia Pestis
Organism: Yersinia pestis
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2017-04-05 Classification: Structural genomics, unknown function Ligands: GOL |
 |
Organism: Staphylococcus epidermidis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2013-09-04 Classification: HYDROLASE |
 |
Crystal Structure Of Bcpa*(D312A), The Major Pilin Subunit Of Bacillus Cereus
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2012-05-30 Classification: CELL ADHESION |
 |
The Structure Of The Slh Domain From B. Anthracis Surface Array Protein At 1.8A
Organism: Bacillus anthracis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2011-04-27 Classification: STRUCTURAL PROTEIN Ligands: SO4 |
 |
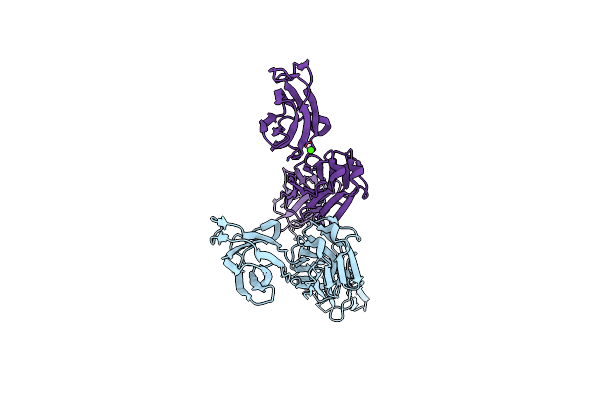
Organism: Oxyuranus microlepidotus
Method: X-RAY DIFFRACTION Resolution:1.33 Å Release Date: 2010-09-08 Classification: ANTIBIOTIC |
 |
Organism: Bacillus cereus atcc 14579
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2009-11-24 Classification: CELL ADHESION Ligands: CA |
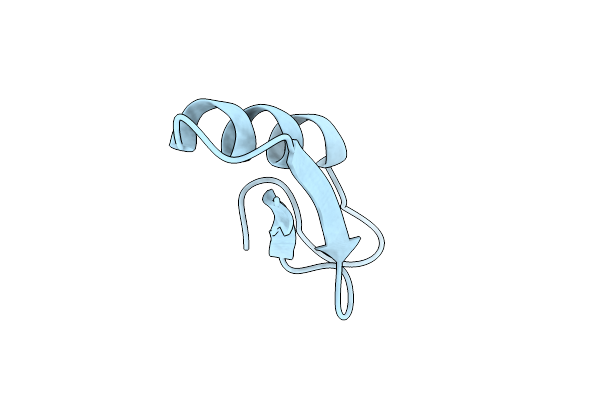 |
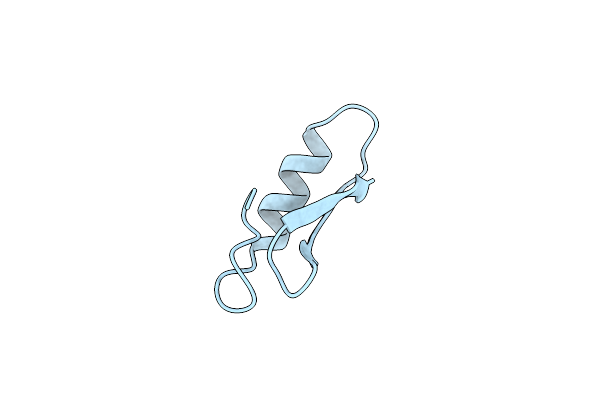
Method: X-RAY DIFFRACTION
Resolution:1.00 Å Release Date: 2009-06-09 Classification: ANTIMICROBIAL PROTEIN |
 |
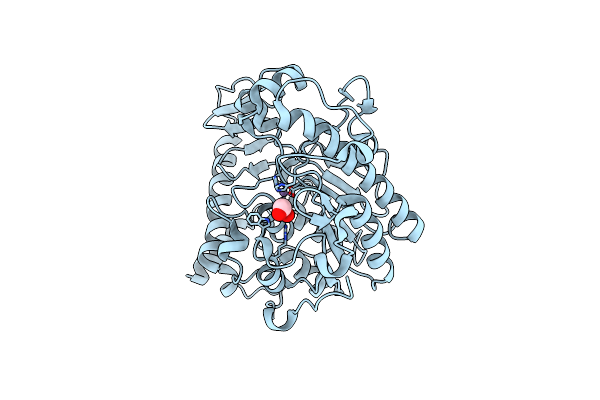
Method: X-RAY DIFFRACTION
Resolution:1.35 Å Release Date: 2009-06-09 Classification: ANTIMICROBIAL PROTEIN |
 |
Structure-Based Mechanism Of Lipoteichoic Acid Synthesis By Staphylococcus Aureus Ltas.
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2009-02-03 Classification: TRANSFERASE Ligands: MN, EDO |

