Search Count: 8
 |
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Resolution:3.40 Å Release Date: 2014-10-08 Classification: RIBOSOME Ligands: ZN, MG |
 |
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Resolution:3.40 Å Release Date: 2014-10-08 Classification: RIBOSOME Ligands: ZN |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2013-11-20 Classification: NUCLEAR PROTEIN/RNA Ligands: ALF, MG, GDP |
 |
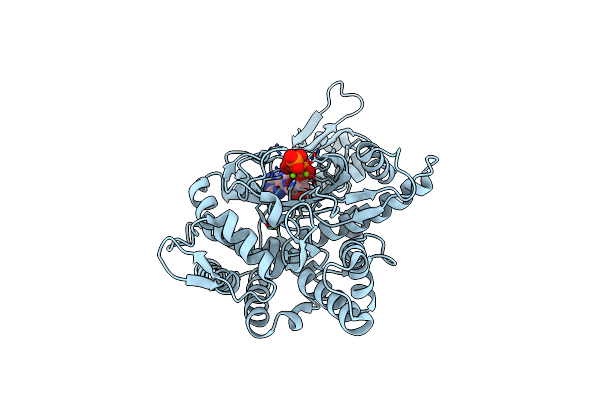
Structure Of The Deamidase-Depupylase Dop Of The Prokaryotic Ubiquitin-Like Modification Pathway
Organism: Acidothermus cellulolyticus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2012-09-12 Classification: HYDROLASE |
 |
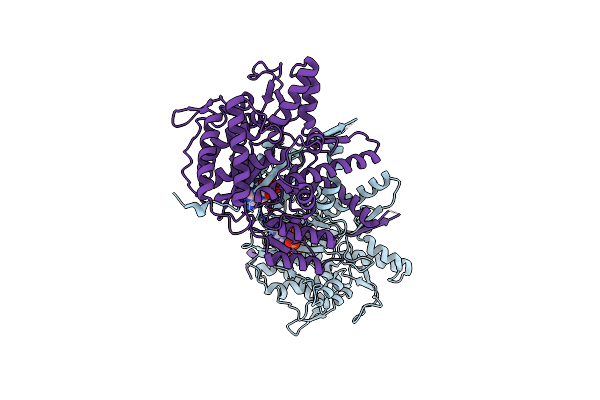
Structure Of The Deamidase-Depupylase Dop Of The Prokaryotic Ubiquitin-Like Modification Pathway In Complex With Atp
Organism: Acidothermus cellulolyticus
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2012-09-12 Classification: HYDROLASE Ligands: ATP, MG |
 |
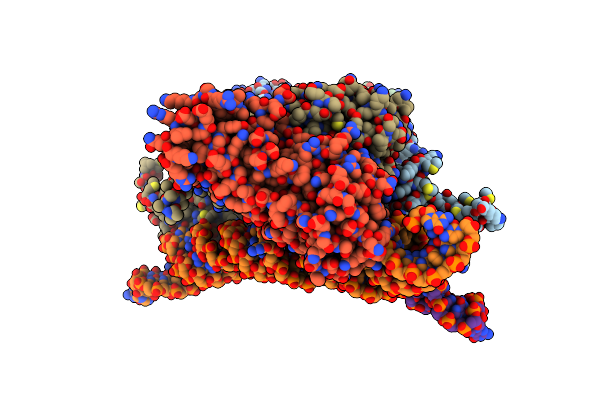
Structure Of The Pup Ligase Pafa Of The Prokaryotic Ubiquitin-Like Modification Pathway In Complex With Adp
Organism: Corynebacterium glutamicum
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2012-09-12 Classification: LIGASE Ligands: ADP, MG |
 |
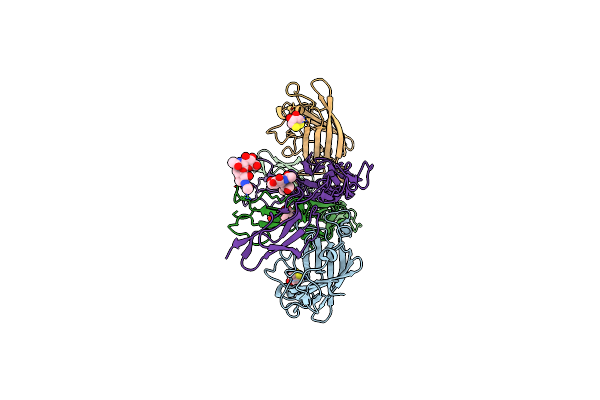
The Crystal Structure Of The Signal Recognition Particle (Srp) In Complex With Its Receptor(Sr)
Organism: Escherichia coli k-12, Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:3.94 Å Release Date: 2011-03-02 Classification: PROTEIN TRANSPORT Ligands: GCP, MG |
 |
Structure Of Extracelllar Portion Of Cd46 In Complex With Adenovirus Type 11 Knob
Organism: Human adenovirus 11, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.84 Å Release Date: 2010-10-13 Classification: CELL ADHESION/IMMUNE SYSTEM Ligands: DTD |

