Search Count: 31
 |
Organism: Scytonema hofmannii, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-09-17 Classification: DNA BINDING PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.90 Å Release Date: 2024-10-16 Classification: TRANSCRIPTION Ligands: RMF |
 |
Organism: Homo sapiens, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2024-03-06 Classification: TRANSCRIPTION Ligands: EDO |
 |
F420-2/Gtp(Gdp) Complex Of F420-Gamma Glutamyl Ligase (Cofe) From Archaeoglobus Fulgidus
Organism: Archaeoglobus fulgidus dsm 4304
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2024-02-28 Classification: LIGASE Ligands: F42, GTP, GDP, MN, NA, SO4 |
 |
Crystal Structure Of Human Cdk12/Cyclin K In Complex With Inhibitor Sr-4835
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.68 Å Release Date: 2023-11-22 Classification: TRANSCRIPTION Ligands: RMF |
 |
Gtp Complex Of F420-Gamma Glutamyl Ligase (Cofe) From Archaeoglobus Fulgidus
Organism: Archaeoglobus fulgidus dsm 4304
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2023-04-12 Classification: LIGASE Ligands: CO3, MN, NA, GTP, SO4 |
 |
F420-1/Gdp Complex Of F420-Gamma Glutamyl Ligase (Cofe) From Archaeoglobus Fulgidus
Organism: Archaeoglobus fulgidus dsm 4304
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-04-12 Classification: LIGASE Ligands: GDP, F4I, MN, NA |
 |
L-Glutamate/Gtp Complex Of F420-Gamma Glutamyl Ligase (Cofe) From Archaeoglobus Fulgidus
Organism: Archaeoglobus fulgidus dsm 4304
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2023-04-12 Classification: LIGASE Ligands: GGL, GTP, MN |
 |
Organism: Scytonema hofmannii, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-12-28 Classification: DNA BINDING PROTEIN Ligands: ATP, MG, ZN |
 |
Organism: Scytonema hofmannii, Escherichia coli k-12, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-12-28 Classification: RNA BINDING PROTEIN Ligands: ATP, MG, ZN |
 |
Organism: Scytonema hofmannii, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-12-28 Classification: RNA BINDING PROTEIN Ligands: ATP, MG |
 |
Organism: Scytonema hofmannii
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2021-12-01 Classification: DNA BINDING PROTEIN Ligands: ZN |
 |
Organism: Scytonema hofmannii, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2021-12-01 Classification: DNA BINDING PROTEIN |
 |
Organism: Scytonema hofmannii, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2021-12-01 Classification: DNA BINDING PROTEIN Ligands: ANP, MG |
 |
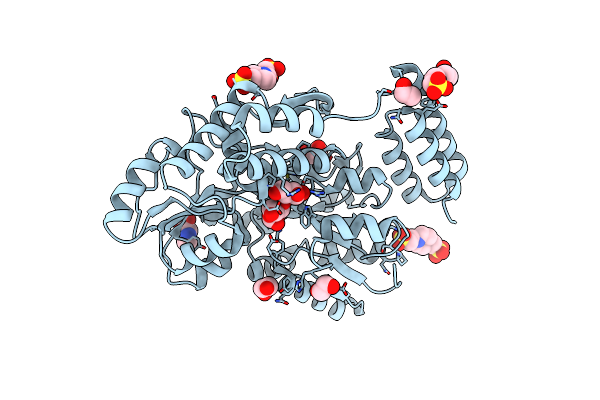
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.64 Å Release Date: 2021-10-27 Classification: IMMUNE SYSTEM |
 |
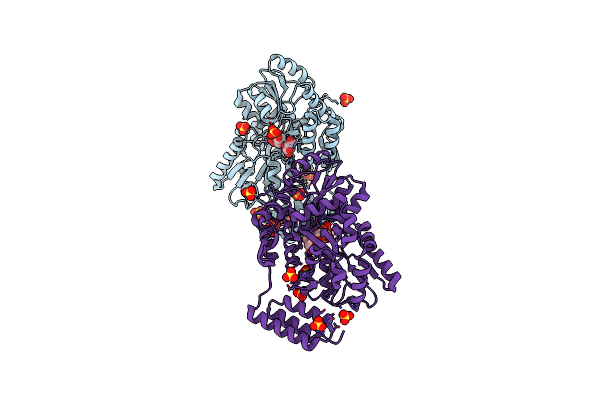
Structure Of The C-Terminal Domain Of The Menangle Virus Phosphoprotein (Residues 329 -388), Fused To Mbp. Space Group P212121
Organism: Serratia sp. (strain fs14), Menangle virus
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2021-10-06 Classification: VIRAL PROTEIN Ligands: EDO, PIN, PRO |
 |
Structure Of The C-Terminal Domain Of The Menangle Virus Phosphoprotein (Residues 329 -388), Fused To Mbp. Space Group P21.
Organism: Serratia sp. (strain fs14), Menangle virus
Method: X-RAY DIFFRACTION Resolution:1.31 Å Release Date: 2021-09-15 Classification: VIRAL PROTEIN Ligands: SO4 |
 |
 |
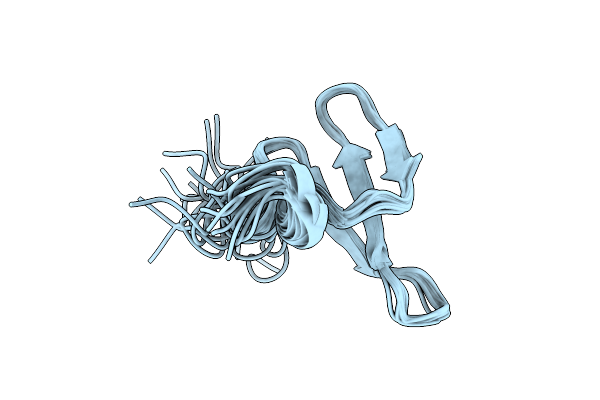
Solution Structure Of Venturia Inaequalis Cellophane-Induced 1 Protein (Vicin1) Domains 1 And 2
Organism: Venturia inaequalis
Method: SOLUTION NMR Release Date: 2012-07-18 Classification: CELL ADHESION |
 |
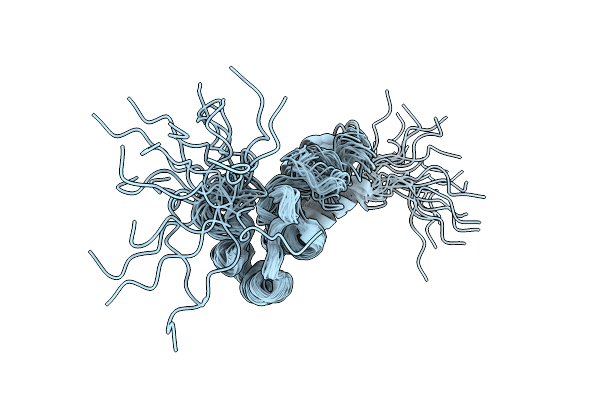
Solution Structure Of E.Coli Rnase P Rna P4 Stem Oligoribonucleotide, U69A Mutation
|

