Search Count: 47
 |
Structure Of Madb, A Class I Dehydrates From Clostridium Maddingley In The Apo State
Organism: Clostridium sp. maddingley mbc34-26
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: BIOSYNTHETIC PROTEIN |
 |
Structure Of Madb, A Class I Dehydrates From Clostridium Maddingley, In Complex With Its Substrate
Organism: Clostridium sp. maddingley mbc34-26
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: BIOSYNTHETIC PROTEIN |
 |
Crystal Structure Hr1 Domain Of Rho-Associated Coiled-Coil Protein Kinases (Rock-Hr1)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-06-21 Classification: SIGNALING PROTEIN Ligands: DTT |
 |
The Structure Of Madc From Clostridium Maddingley Reveals New Insights Into Class I Lanthipeptide Cyclases
Organism: Clostridium
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-02-22 Classification: BIOSYNTHETIC PROTEIN Ligands: ZN, NA, MPD, ACT, CL |
 |
Organism: Saccharomyces
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2022-08-17 Classification: TRANSCRIPTION |
 |
Organism: Ustilago maydis
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2022-06-15 Classification: RNA BINDING PROTEIN |
 |
Cryo-Em Structure Of Pdr5 From Saccharomyces Cerevisiae In Inward-Facing Conformation Without Nucleotides
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2021-11-10 Classification: TRANSPORT PROTEIN |
 |
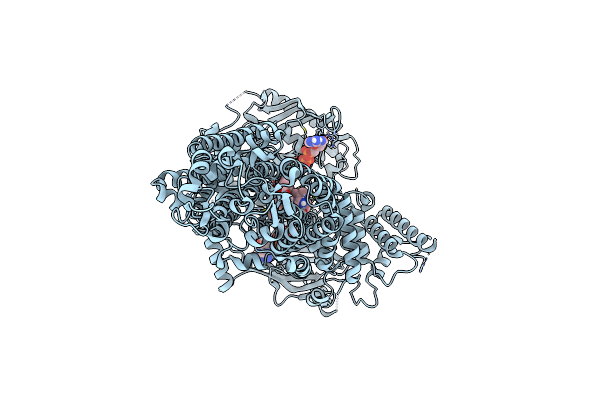
Cryo-Em Structure Of Pdr5 From Saccharomyces Cerevisiae In Inward-Facing Conformation With Adp/Atp
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2021-11-10 Classification: TRANSPORT PROTEIN Ligands: ATP, ADP |
 |
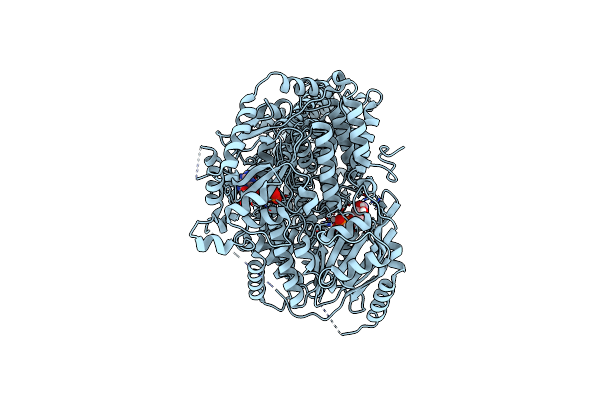
Cryo-Em Structure Of Pdr5 From Saccharomyces Cerevisiae In Inward-Facing Conformation With Adp/Atp And Rhodamine 6G
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2021-11-10 Classification: TRANSPORT PROTEIN Ligands: ATP, ADP, RHQ |
 |
Cryo-Em Structure Of Pdr5 From Saccharomyces Cerevisiae In Outward-Facing Conformation With Adp-Orthovanadate/Atp
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2021-11-10 Classification: TRANSPORT PROTEIN Ligands: ATP, MG, AOV |
 |
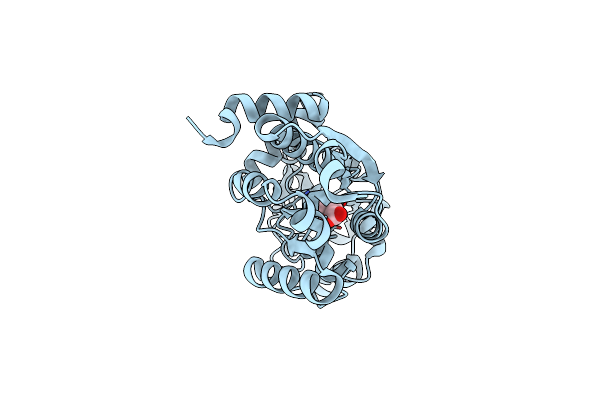
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2018-11-21 Classification: TRANSPORT PROTEIN Ligands: BET |
 |
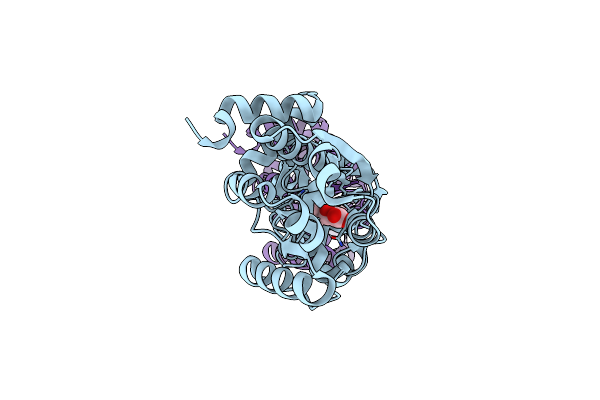
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2018-11-21 Classification: TRANSPORT PROTEIN Ligands: DQY |
 |
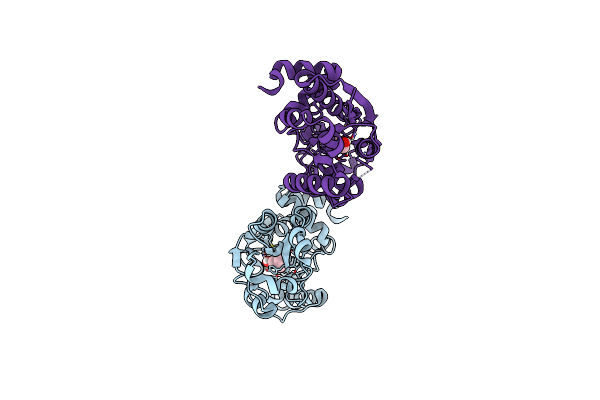
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2018-11-21 Classification: TRANSPORT PROTEIN Ligands: 152 |
 |
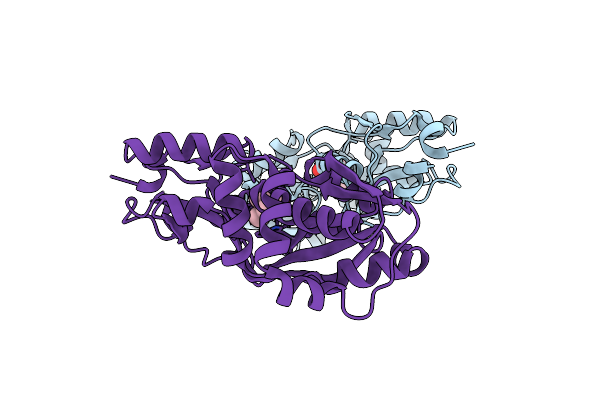
Organism: Bacillus subtilis (strain 168)
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2018-11-21 Classification: TRANSPORT PROTEIN Ligands: CHT |
 |
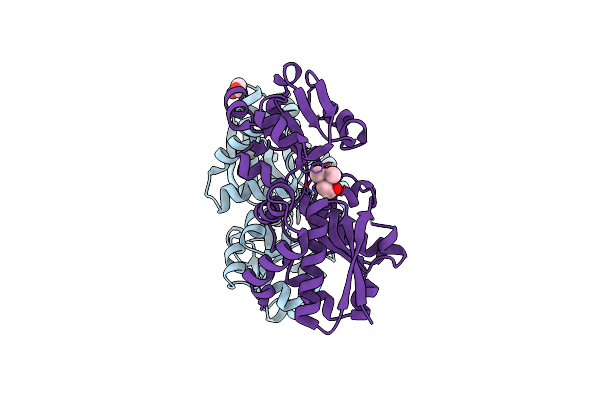
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2018-03-28 Classification: TRANSPORT PROTEIN Ligands: 3Q7 |
 |
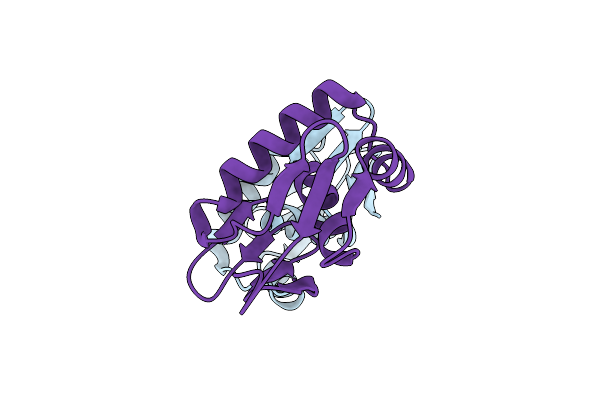
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-03-14 Classification: CHOLINE-BINDING PROTEIN Ligands: 1Y8, EDO |
 |
Structure Of A Lantibiotic Response Regulator: C-Terminal Domain Of The Nisin Resistance Regulator Nsrr
Organism: Streptococcus agalactiae
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2016-07-06 Classification: SIGNALING PROTEIN |
 |
Structure Of A Lantibiotic Response Regulator: N Terminal Domain Of The Nisin Resistance Regulator Nsrr
Organism: Streptococcus agalactiae
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2016-03-16 Classification: SIGNALING PROTEIN Ligands: EDO |
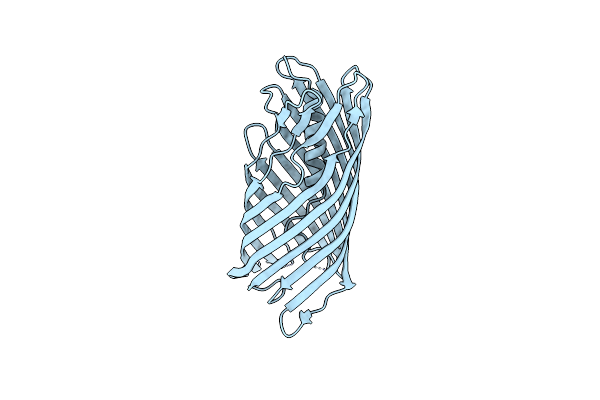 |
Crystal Structure Of The Transport Unit Of The Autotransporter Aida-I From Escherichia Coli
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2014-06-04 Classification: PROTEIN BINDING |
 |

