Search Count: 502
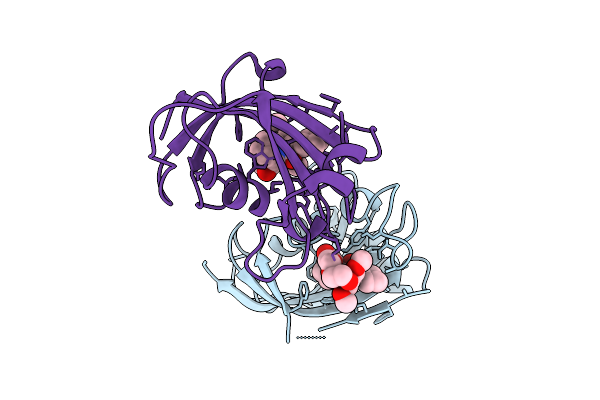 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: ISOMERASE Ligands: A1I5X |
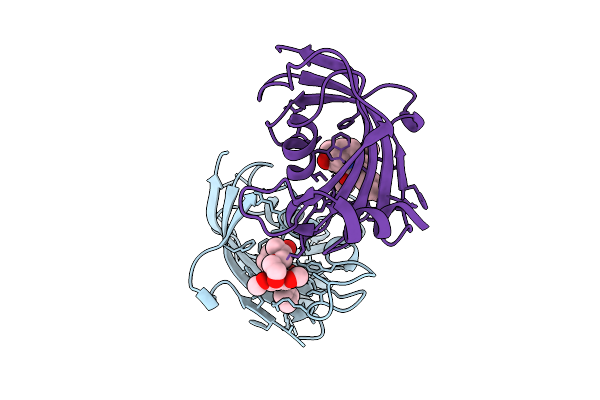 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: ISOMERASE Ligands: A1I53 |
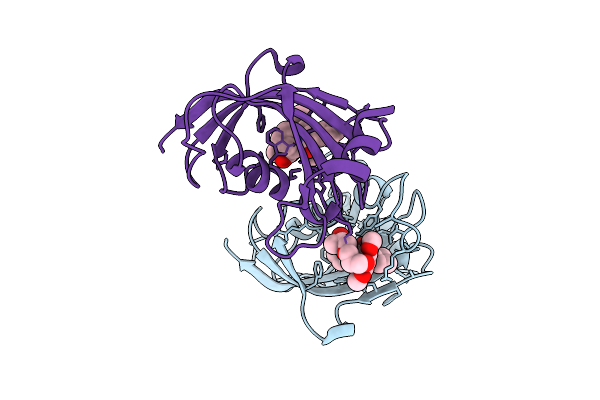 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: ISOMERASE Ligands: A1I54 |
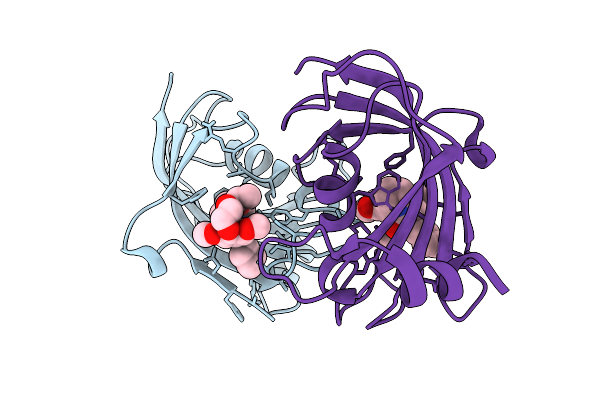 |
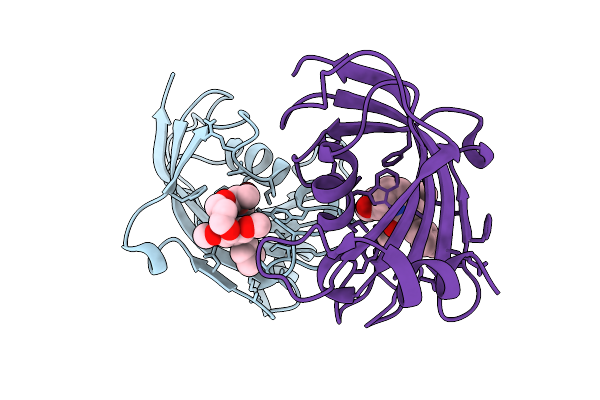
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: ISOMERASE Ligands: A1I55 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: ISOMERASE Ligands: A1I5Y |
 |
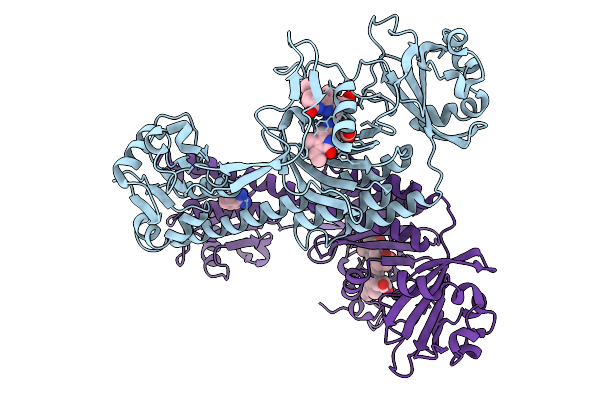
Structure Of The 70S-Ef-G(P610L)-Gdp-Pi Ribosome Complex With Trnas In Hybrid State 2 (H2-Ef-G(P610L)-Gdp-Pi)
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: RIBOSOME Ligands: MG, ZN, NA, AM2, GDP, PO4 |
 |
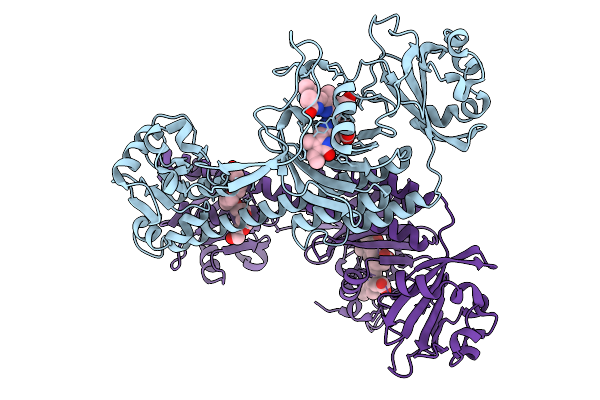
Photoactivation In Bacteriophytochromes, Reference (Dark) Structure For The 3 Ps Time Point
Organism: Stigmatella aurantiaca
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: SIGNALING PROTEIN Ligands: 3Q8, BEN |
 |
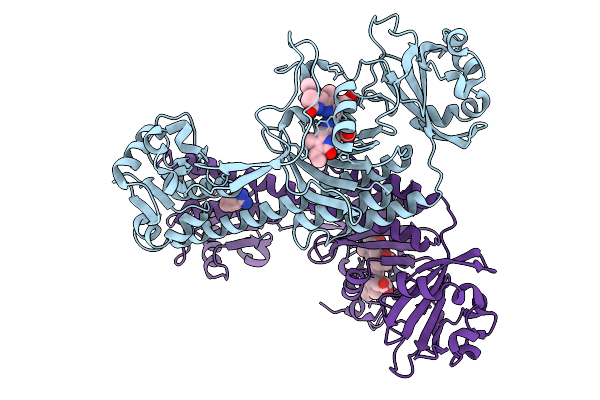
Photoactivation In Bacteriophytochrome, High Resolution Cryo Structure In The Dark.
Organism: Stigmatella aurantiaca
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: SIGNALING PROTEIN Ligands: EL5, P33 |
 |
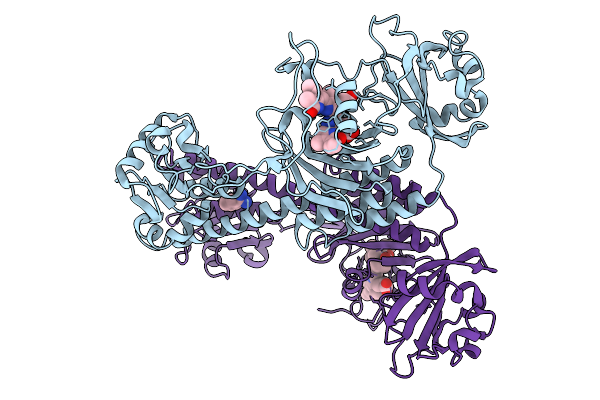
Photoactivation In Bacteriophytochromes, Reference (Dark) Structure For The 100 Ps Time Point
Organism: Stigmatella aurantiaca
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: SIGNALING PROTEIN Ligands: EL5, BEN |
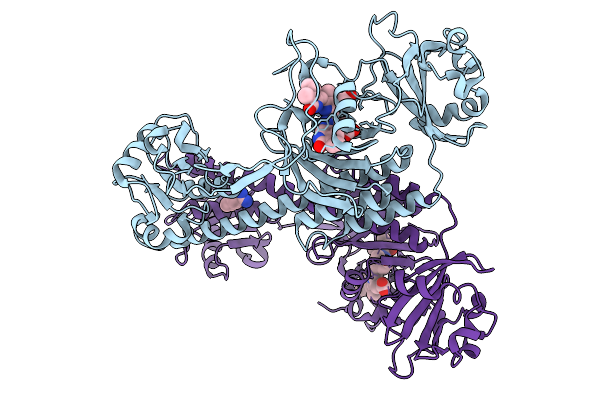
 |
Organism: Stigmatella aurantiaca
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: SIGNALING PROTEIN Ligands: BLA, BEN |
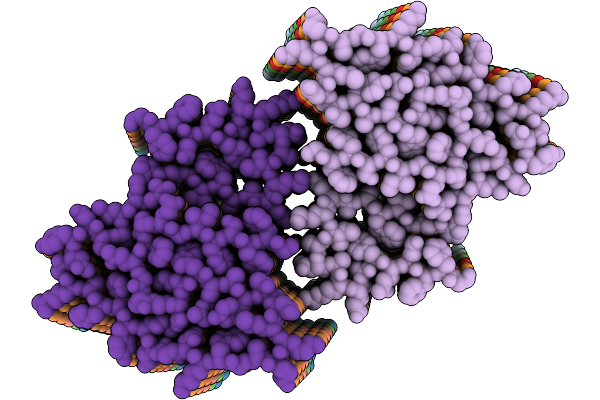
 |
Organism: Stigmatella aurantiaca
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: SIGNALING PROTEIN Ligands: BLA, BEN |
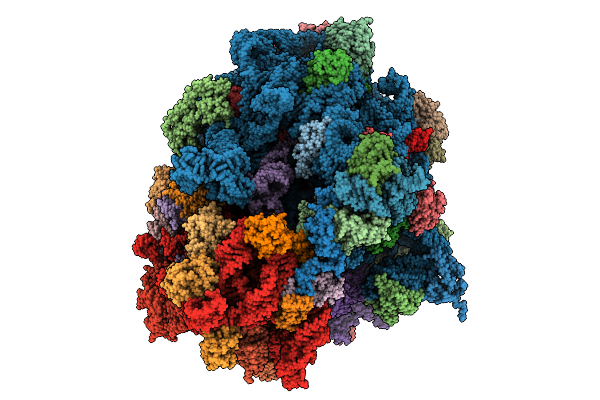
 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: PROTEIN FIBRIL |
 |
Structure Of The 70S-Ef-G(P610L)-Gdp-Pi Ribosome Complex With Trnas In Hybrid State 1 (H1-Ef-G(P610L)-Gdp-Pi)
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: RIBOSOME Ligands: MG, ZN, NA, AM2, PO4, GDP |
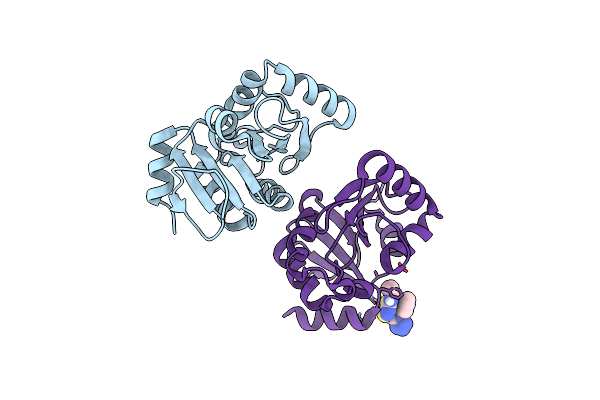 |
Pandda Analysis Group Deposition -- Crystal Structure Of Sars-Cov-2 Nsp3 Macrodomain In Complex With Avi-0000286
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: VIRAL PROTEIN, Hydrolase Ligands: A1A2H |
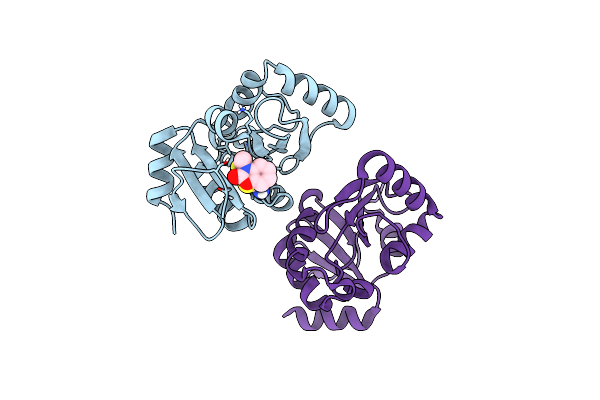 |
Pandda Analysis Group Deposition -- Crystal Structure Of Sars-Cov-2 Nsp3 Macrodomain In Complex With Avi-0000296
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: VIRAL PROTEIN, Hydrolase Ligands: A1A41 |
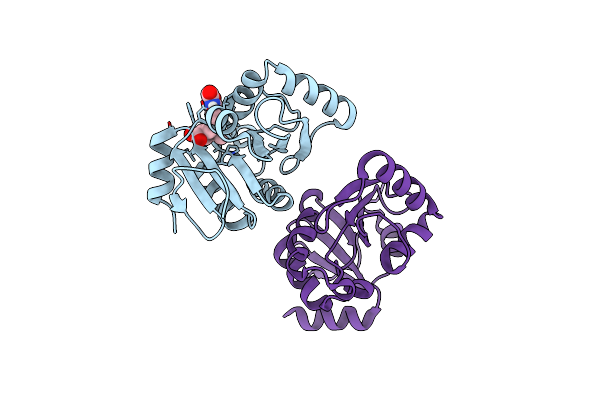 |
Pandda Analysis Group Deposition -- Crystal Structure Of Sars-Cov-2 Nsp3 Macrodomain In Complex With Avi-0000303
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: VIRAL PROTEIN, Hydrolase Ligands: A1A42 |
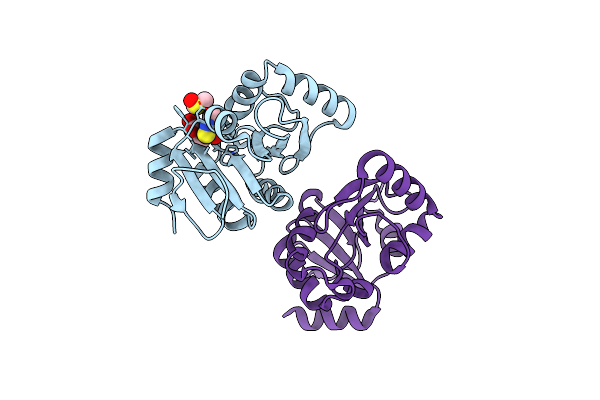 |
Pandda Analysis Group Deposition -- Crystal Structure Of Sars-Cov-2 Nsp3 Macrodomain In Complex With Avi-0000002
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: VIRAL PROTEIN, Hydrolase Ligands: DMS, A1A47 |
 |
Pandda Analysis Group Deposition -- Crystal Structure Of Sars-Cov-2 Nsp3 Macrodomain In Complex With Avi-0000313
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: VIRAL PROTEIN, Hydrolase Ligands: A1A49, A1A5A, CL |
 |
Pandda Analysis Group Deposition -- Crystal Structure Of Sars-Cov-2 Nsp3 Macrodomain In Complex With Avi-0000316
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: VIRAL PROTEIN, Hydrolase Ligands: A1A5B |
 |
Pandda Analysis Group Deposition -- Crystal Structure Of Sars-Cov-2 Nsp3 Macrodomain In Complex With Avi-0000317
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: VIRAL PROTEIN, Hydrolase Ligands: A1A5C |

