Search Count: 28
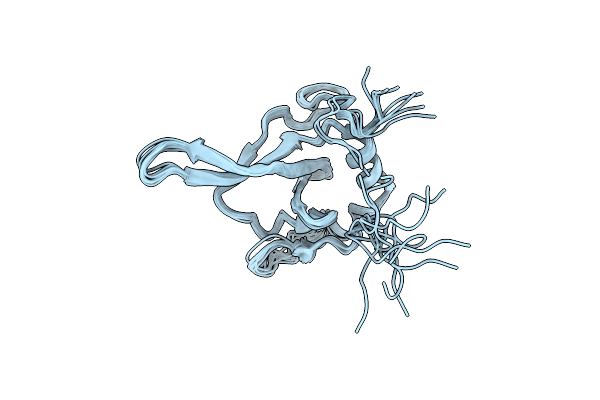 |
Organism: Enterobacteria phage fd
Method: SOLUTION NMR Release Date: 2025-06-04 Classification: VIRAL PROTEIN |
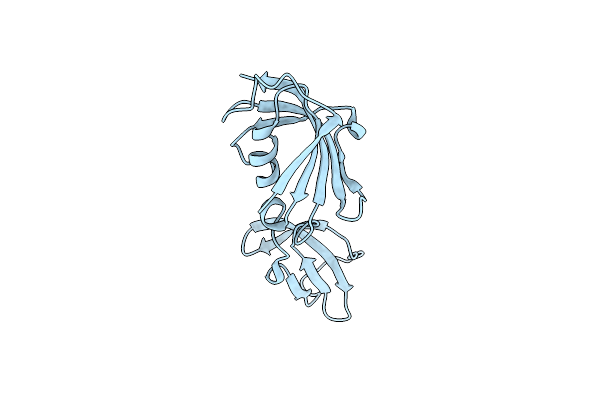 |
Crystal Structure Of Fkbp12-If(Slyd), A Chimeric Protein Of Human Fkbp12 And The Insert In Flap Domain Of Ecoli Slyd
Organism: Homo sapiens, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-03-08 Classification: ISOMERASE |
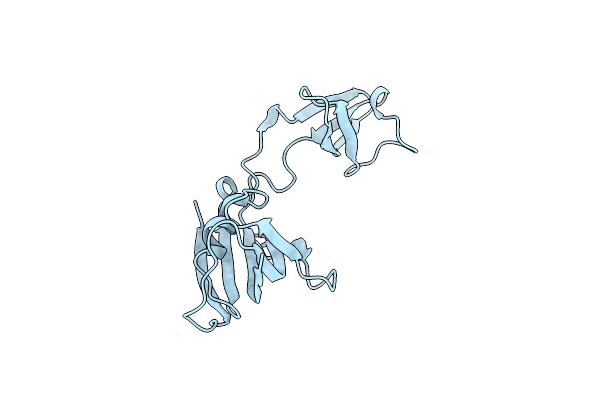 |
Crystal Structure Of Fkbp12-If(Slpa), A Chimeric Protein Of Human Fkbp12 And The Insert In Flap Domain Of Ecoli Slpa
Organism: Homo sapiens, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2017-03-08 Classification: ISOMERASE |
 |
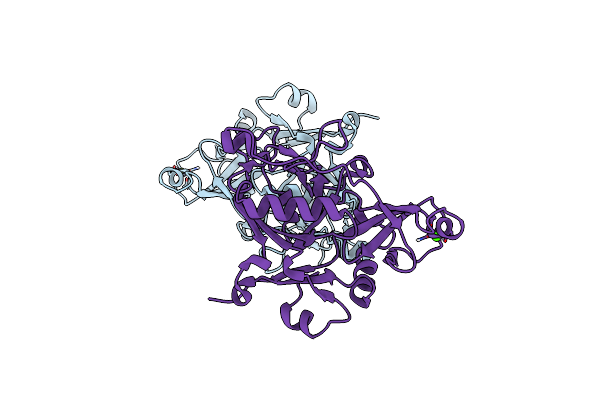
Organism: Hirschia baltica (strain atcc 49814 / dsm 5838 / ifam 1418)
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2016-06-22 Classification: ISOMERASE Ligands: MG |
 |
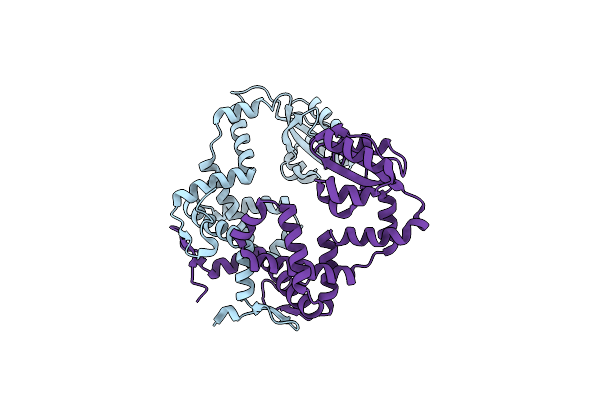
Organism: Hirschia baltica
Method: X-RAY DIFFRACTION Resolution:1.29 Å Release Date: 2016-06-22 Classification: ISOMERASE Ligands: MG, CA, CL |
 |
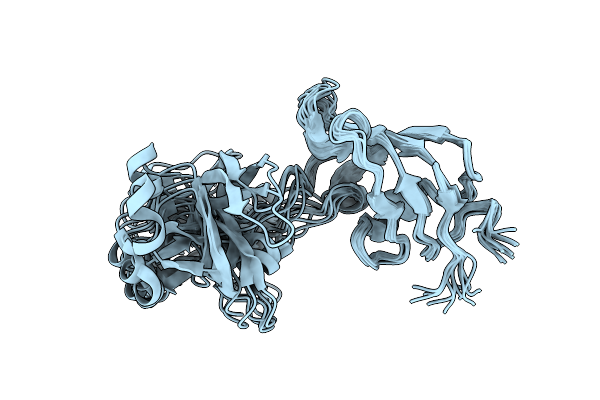
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.63 Å Release Date: 2014-12-24 Classification: ISOMERASE |
 |
 |
Organism: Enterobacteria phage ike
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2012-05-30 Classification: VIRAL PROTEIN |
 |
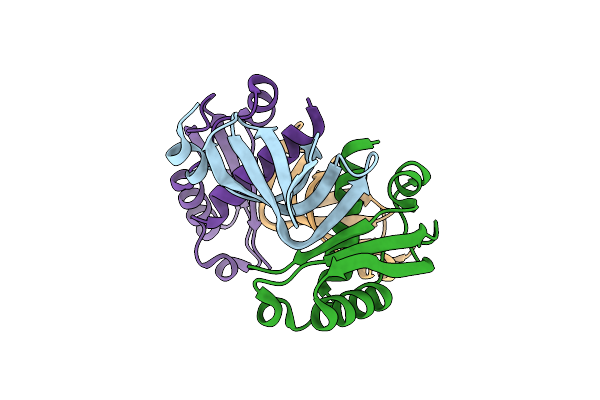
Crystal Structure Of The Tola Binding Domain From The Filamentous Phage Ike
Organism: Enterobacteria phage ike
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2012-05-30 Classification: VIRAL PROTEIN Ligands: MG |
 |
Crystal Structure Of G3P From Phage If1 In Complex With Its Coreceptor, The C-Terminal Domain Of Tola
Organism: Enterobacteria phage if1, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.47 Å Release Date: 2010-12-01 Classification: VIRAL PROTEIN |
 |
Organism: Enterobacteria phage if1
Method: X-RAY DIFFRACTION Resolution:2.92 Å Release Date: 2010-12-01 Classification: VIRAL PROTEIN |
 |
 |
Organism: Enterobacteria phage fd
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2009-11-24 Classification: VIRAL PROTEIN |
 |
Structural And Energetic Determinants For Hyperstable Variants Of Gb1 Obtained From In-Vitro Evolution
Organism: Streptococcus sp. 'group g'
Method: X-RAY DIFFRACTION Resolution:0.88 Å Release Date: 2009-08-18 Classification: PROTEIN BINDING Ligands: CA |
 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2008-08-05 Classification: HYDROLASE |
 |
Changing The Determinants Of Protein Stability From Covalent To Non-Covalent Interactions By In-Vitro Evolution: A Structural And Energetic Analysis
Organism: Bacteriophage fd
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2008-07-22 Classification: VIRAL PROTEIN |
 |
Organism: Streptococcus sp.
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2008-01-08 Classification: PROTEIN BINDING |
 |
Organism: Streptococcus sp.
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2007-12-04 Classification: PROTEIN BINDING |
 |
Crystal Structure Of Bacillus Subtilis Cold Shock Protein Variant Bs-Cspb M1R/E3K/K65I
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2007-05-22 Classification: GENE REGULATION |

