Search Count: 85
 |
Organism: Segatella buccae, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-02-12 Classification: RNA BINDING PROTEIN |
 |
Crystal Structure Of The Condensation Domain Tombc From The Tomaymycin Non-Ribosomal Peptide Synthetase
Organism: Streptomyces regensis
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2024-06-26 Classification: BIOSYNTHETIC PROTEIN Ligands: FMT, GOL, NA, K |
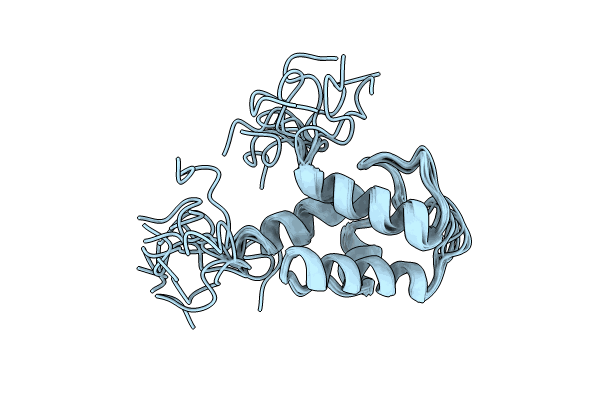 |
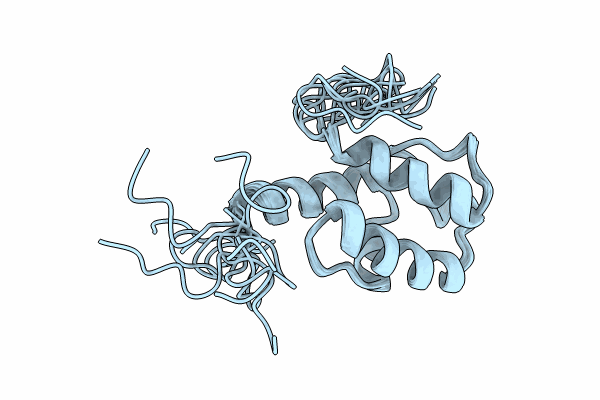
Solution Nmr Structure Of The Peptidyl Carrier Domain Tomapcp From The Tomaymycin Non-Ribosomal Peptide Synthetase
Organism: Streptomyces regensis
Method: SOLUTION NMR Release Date: 2024-06-26 Classification: BIOSYNTHETIC PROTEIN |
 |
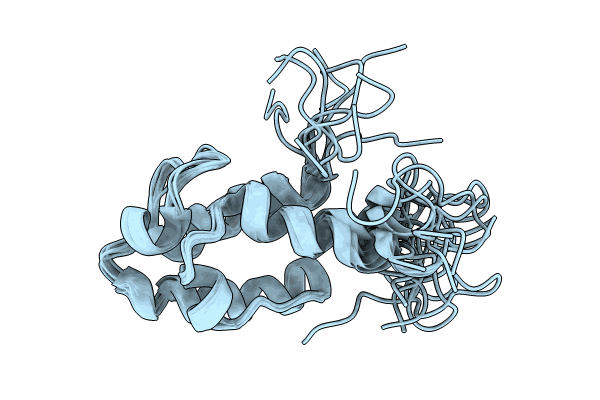
Solution Nmr Structure Of The Peptidyl Carrier Domain Tomapcp From The Tomaymycin Non-Ribosomal Peptide Synthetase In Its Substrate-Loaded State
Organism: Streptomyces regensis
Method: SOLUTION NMR Release Date: 2024-06-26 Classification: BIOSYNTHETIC PROTEIN |
 |
Solution Nmr Structure Of The Novel Adaptor Domain Tombn91 From The Tomaymycin Non-Ribosomal Peptide Synthetase
Organism: Streptomyces regensis
Method: SOLUTION NMR Release Date: 2024-06-26 Classification: BIOSYNTHETIC PROTEIN |
 |
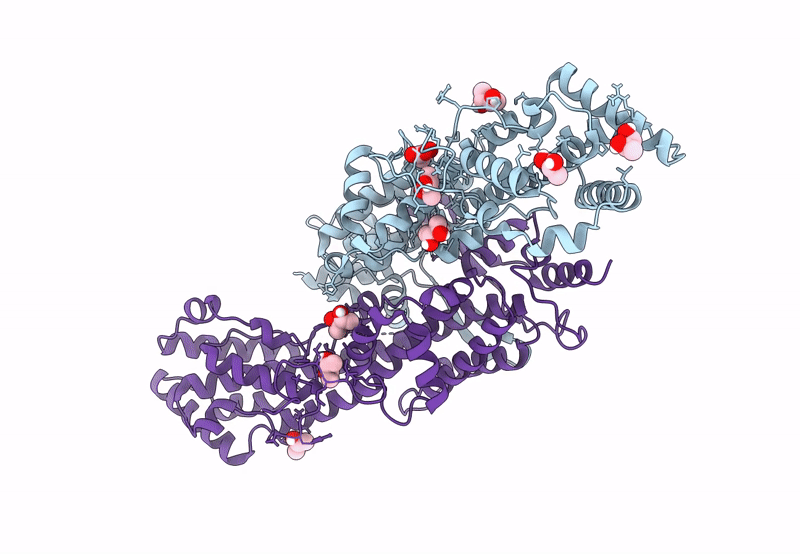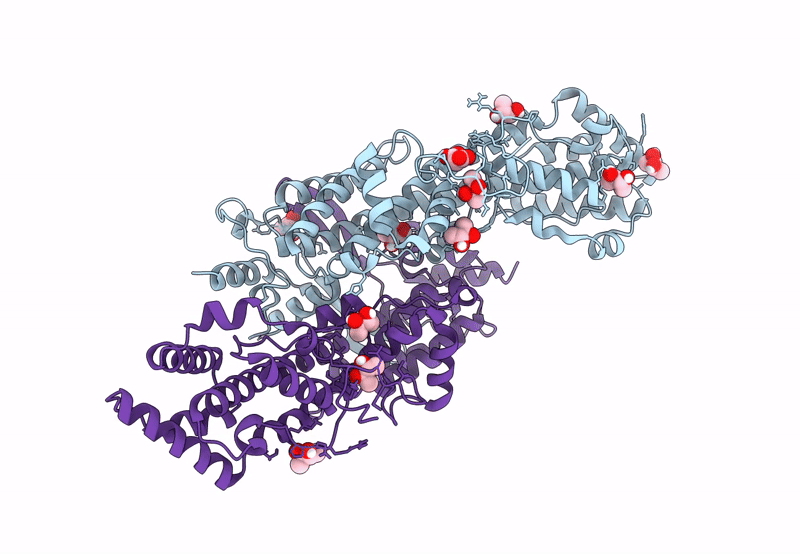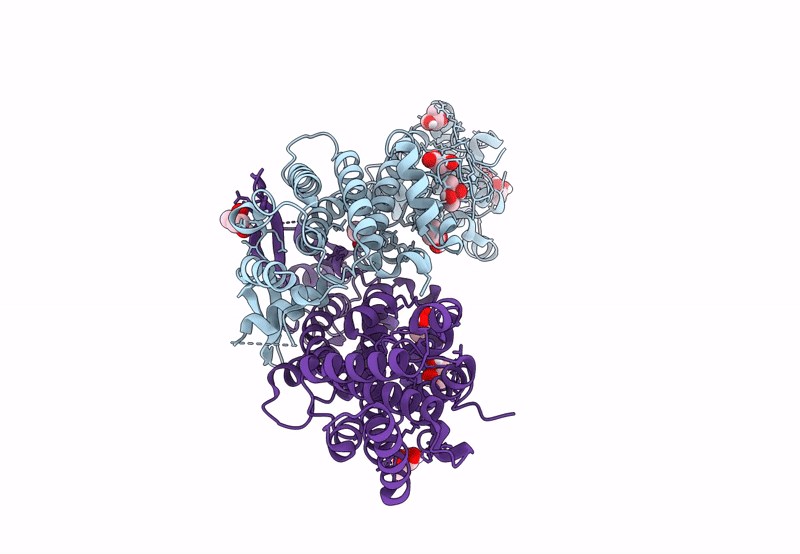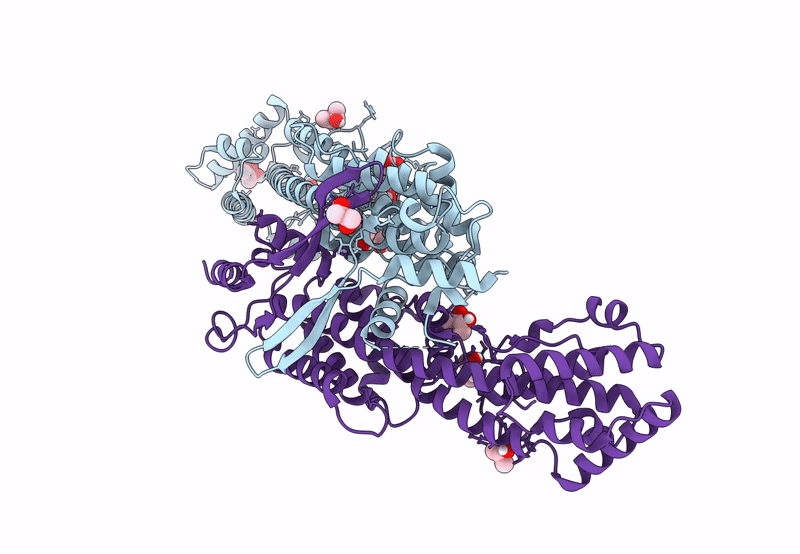
Semet Derivative Structure Of The Condensation Domain Tombc From The Tomaymycin Non-Ribosomal Peptide Synthetase
Organism: Streptomyces regensis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-06-26 Classification: BIOSYNTHETIC PROTEIN Ligands: FMT, K, GOL |
 |
Organism: Muromegalovirus
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2023-10-11 Classification: STRUCTURAL PROTEIN Ligands: BU3, MLI |
 |
Fusion Construct Of Pqse And Rhlr In Complex With The Synthetic Antagonist Mbtl
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:3.46 Å Release Date: 2022-12-14 Classification: TRANSCRIPTION Ligands: FE, K5G |
 |
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2022-12-14 Classification: HYDROLASE Ligands: FE, CAC, BEZ |
 |
Pross Optimitzed Variant Of Rhlr (75 Mutations) In Complex With The Synthetic Antagonist Mbtl
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2022-12-14 Classification: TRANSCRIPTION Ligands: K5G |
 |
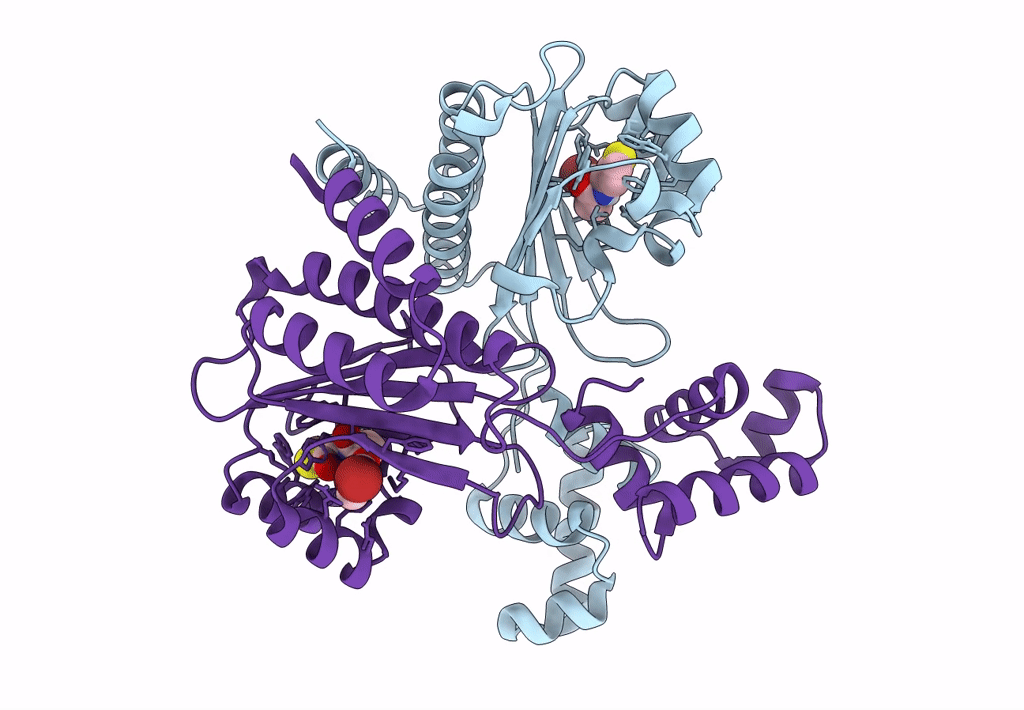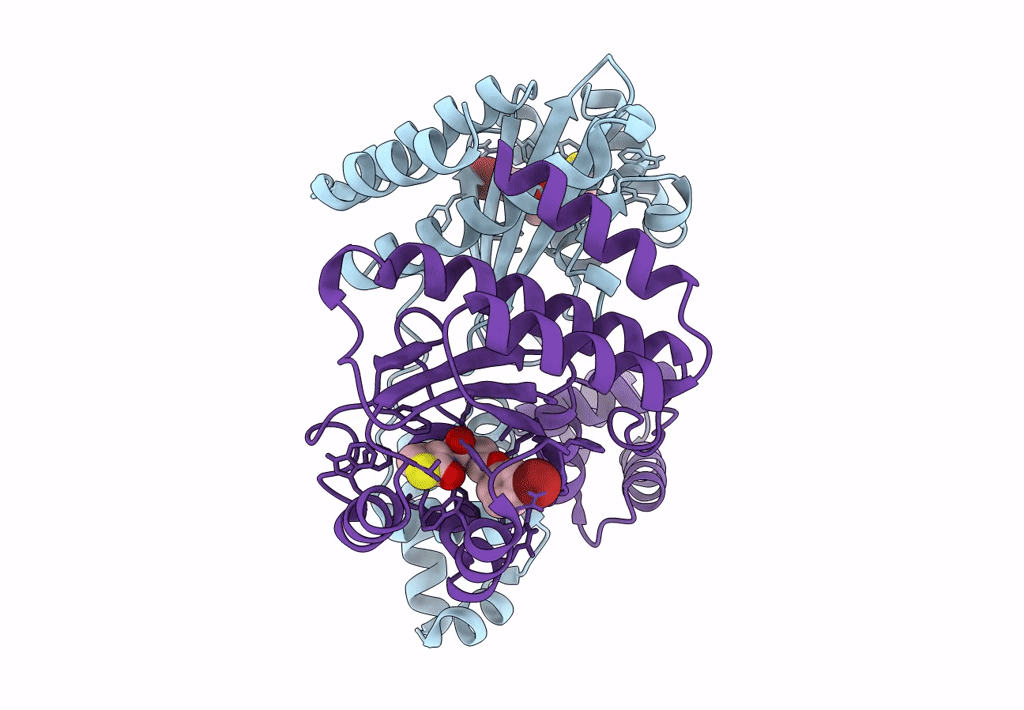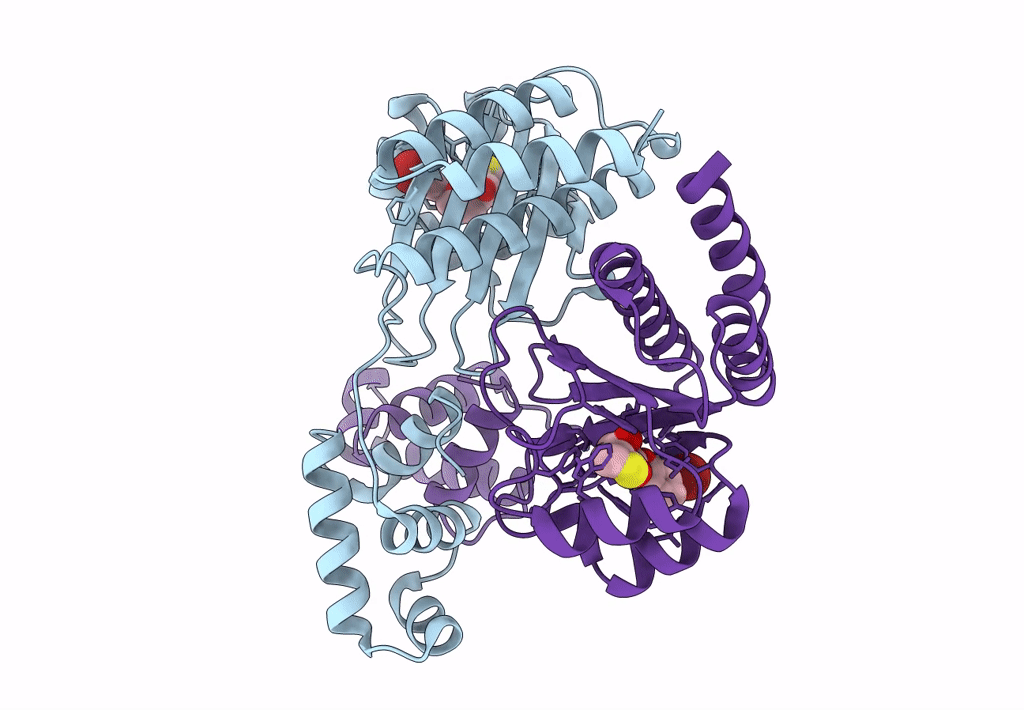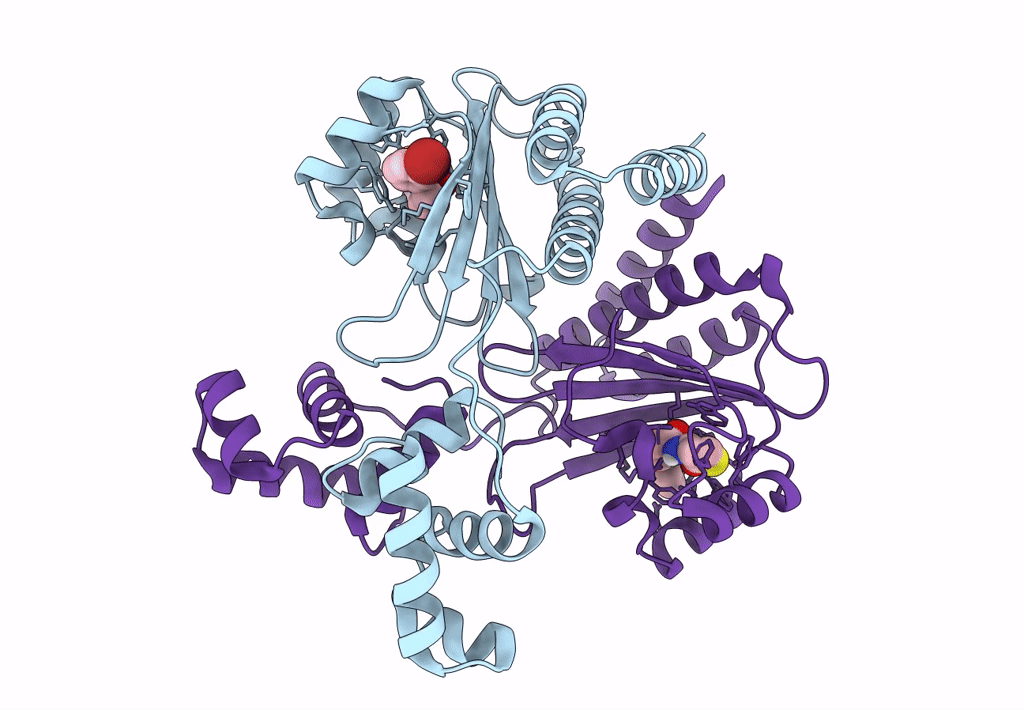
Pross Optimitzed Variant Of Rhlr (75 Mutations) In Complex With Native Autoinducer C4-Hsl
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:3.49 Å Release Date: 2022-12-14 Classification: TRANSCRIPTION Ligands: HL4 |
 |
Pross Optimitzed Variant Of Rhlr (61 Mutations) In Complex With The Synthetic Antagonist Mbtl
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2022-12-14 Classification: TRANSCRIPTION Ligands: K5G |
 |
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:3.06 Å Release Date: 2022-12-14 Classification: TRANSCRIPTION Ligands: FE, K5G |
 |
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:3.06 Å Release Date: 2022-12-14 Classification: SIGNALING PROTEIN Ligands: FE, HL4 |
 |
Crystal Structure Of Pqsr (Mvfr) Ligand-Binding Domain In Complex With Compound N-((2-(4-Cyclopropylphenyl)Thiazol-5-Yl)Methyl)-2-(Trifluoromethyl)Pyridin-4-Amine
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2022-11-30 Classification: GENE REGULATION Ligands: XBH |
 |
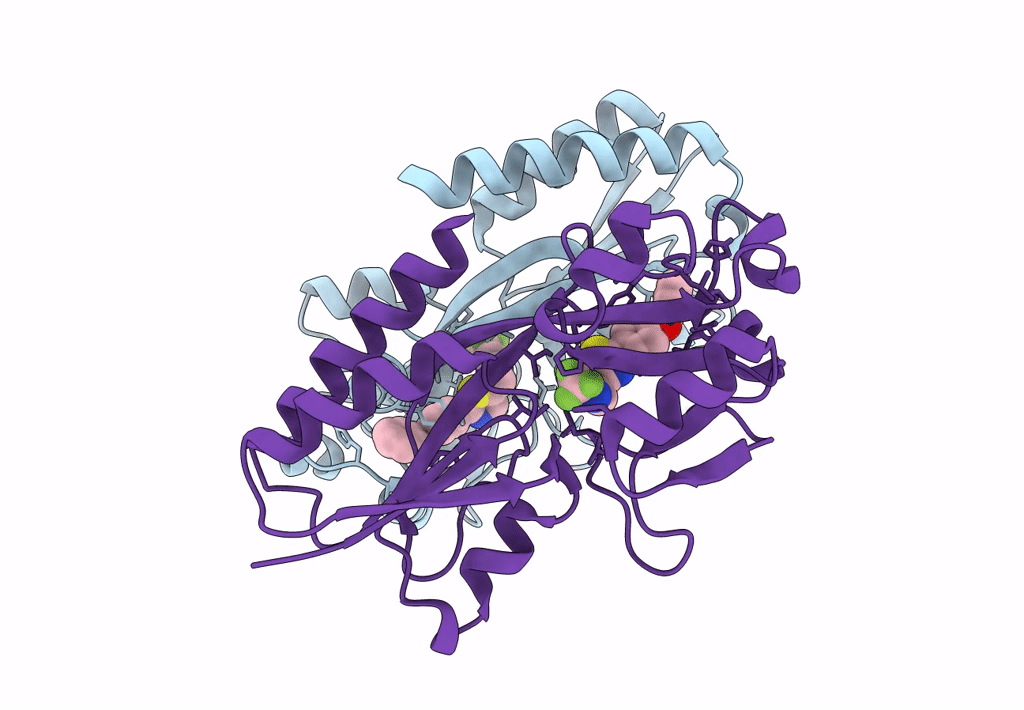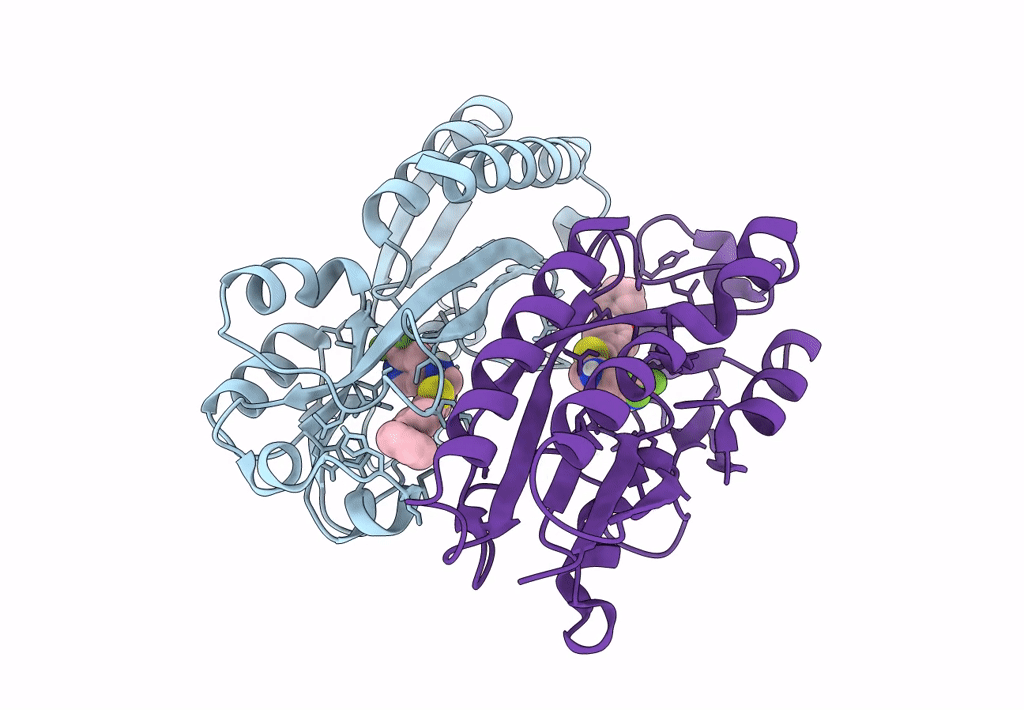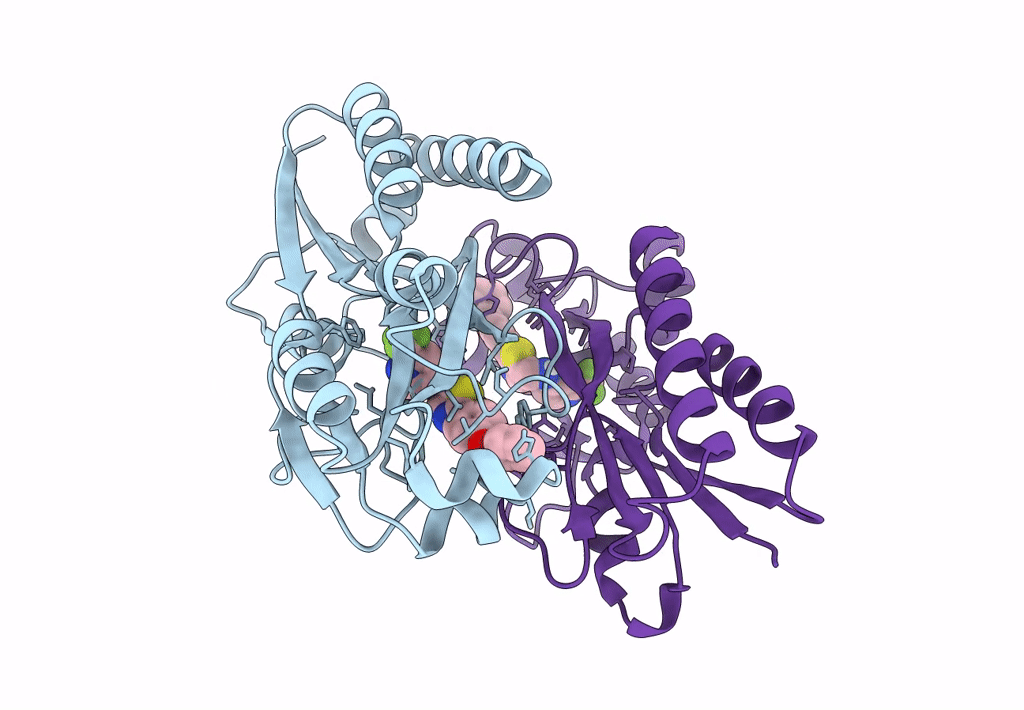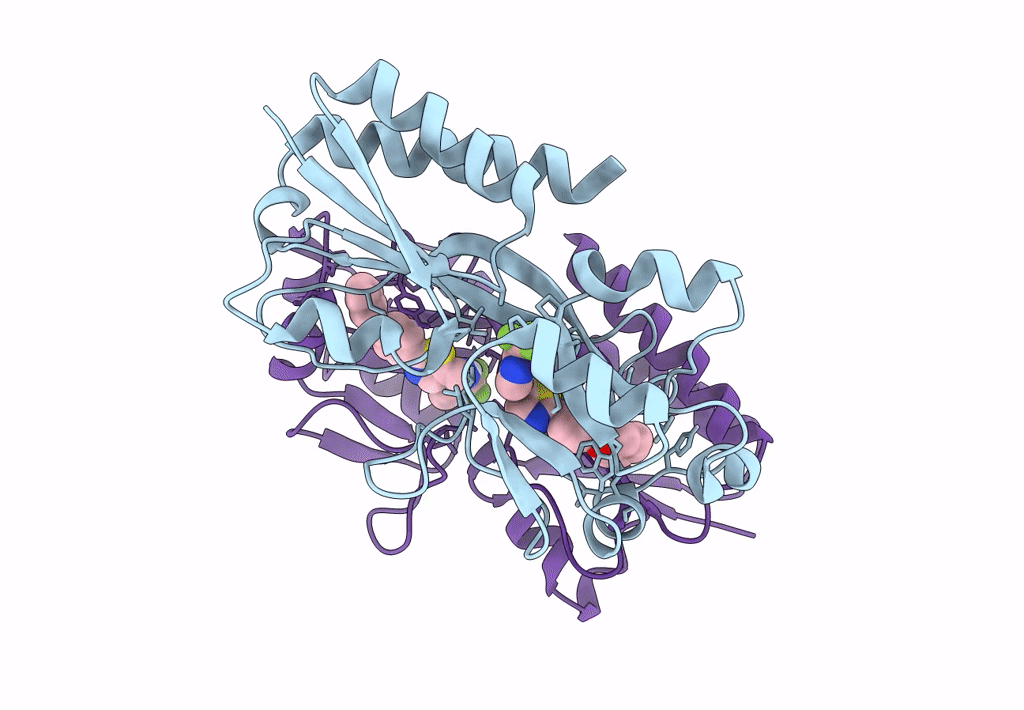
Crystal Structure Of Pqsr (Mvfr) Ligand-Binding Domain In Complex With Compound 1456
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:2.67 Å Release Date: 2022-11-23 Classification: GENE REGULATION Ligands: 9ZL |
 |
Crystal Structure Of Pqsr (Mvfr) Ligand-Binding Domain In Complex With Compound N-((2-(4-Phenoxyphenyl)Thiazol-5-Yl)Methyl)-2-(Trifluoromethyl)Pyridin-4-Amine
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:2.67 Å Release Date: 2022-11-23 Classification: GENE REGULATION Ligands: A0F |
 |
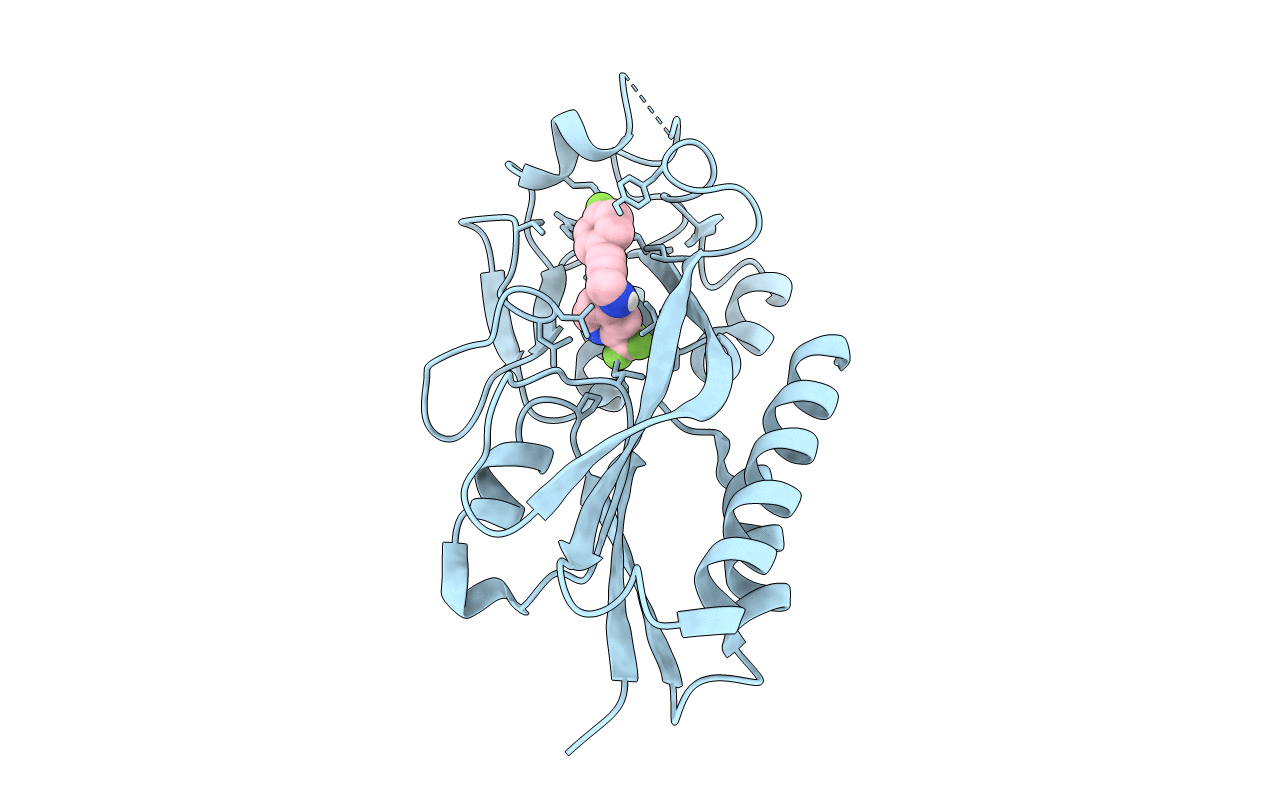
Crystal Structure Of Pqsr (Mvfr) Ligand-Binding Domain In Complex With 3-Pyridin-4-Yl-2,4-Dihydro-Indeno[1,2-.C.]Pyrazole
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:2.74 Å Release Date: 2022-07-27 Classification: GENE REGULATION Ligands: 5N9 |
 |
Crystal Structure Of Pqsr (Mvfr) Ligand-Binding Domain In Complex With A Pyridin Agonist
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2021-10-06 Classification: DNA BINDING PROTEIN Ligands: U7Q |
 |
Crystal Structure Of Pqsr (Mvfr) Ligand-Binding Domain In Complex With Triazolo-Pyridine Inverse Agonist A
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2021-04-14 Classification: DNA BINDING PROTEIN Ligands: OT2, OT8, MG |

