Search Count: 103
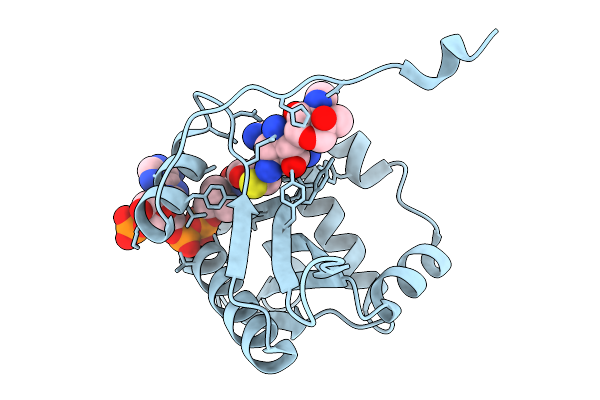 |
Structure Of Aminoglycoside Acetyltransferase Aac(3)-Ia In Complex With Sisomicin And Coa
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: TRANSFERASE Ligands: COA, SIS |
 |
Structure Of N-3 Aminoglycoside Acetyltransferase Xia (Aac(3)-Xia) In Complex With Coenzyme A And Tobramycin
Organism: Corynebacterium striatum
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: TRANSFERASE Ligands: COA, TOY |
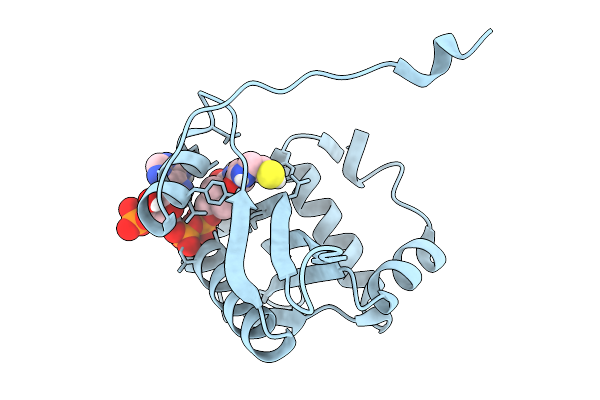 |
Structure Of Aminoglycoside Acetyltransferase Aac(3)-Ia In Complex With Coa
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: TRANSFERASE Ligands: COA |
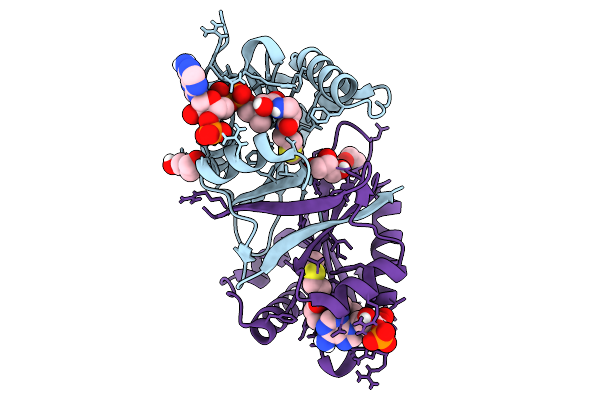 |
Structure Of N-3 Aminoglycoside Acetyltransferase Xi (Aac(3)-Xi) In Complex With Acetyl Coenzyme A
Organism: Corynebacterium striatum
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: TRANSFERASE Ligands: ACO, PEG, GOL |
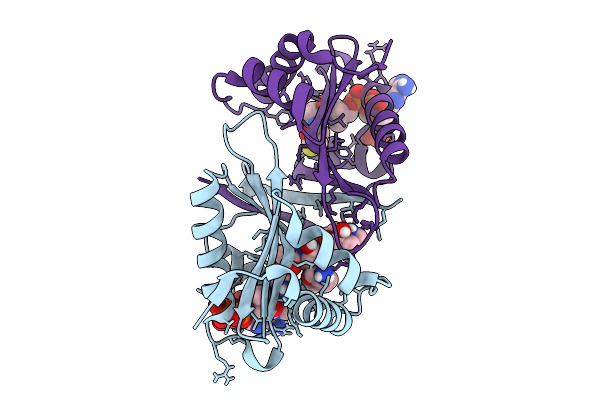 |
Structure Of N-3 Aminoglycoside Acetyltransferase Xi (Aac(3)-Xi) In Complex With Coenzyme A And Acetylated Sisomicin
Organism: Corynebacterium striatum
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: TRANSFERASE Ligands: COA, DVM, ACO |
 |
Structure Of N-3 Aminoglycoside Acetyltransferase Xia (Aac(3)-Xia) In Complex With Acetylamino Coenzyme A And Gentamicin
Organism: Corynebacterium striatum
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: TRANSFERASE Ligands: 51G, 6IM, ACT |
 |
Structure Of N-3 Aminoglycoside Acetyltransferase Xi (Aac(3)-Xi) In Complex With Coenzyme A, And One Apo Monomer
Organism: Corynebacterium striatum
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: TRANSFERASE Ligands: ACO, GOL, PO4 |
 |
Organism: Brevibacillus parabrevis
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2025-03-05 Classification: BIOSYNTHETIC PROTEIN Ligands: J8T |
 |
Organism: Brevibacillus parabrevis
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2024-11-20 Classification: BIOSYNTHETIC PROTEIN Ligands: GOL, JQG, A1AMO, MN |
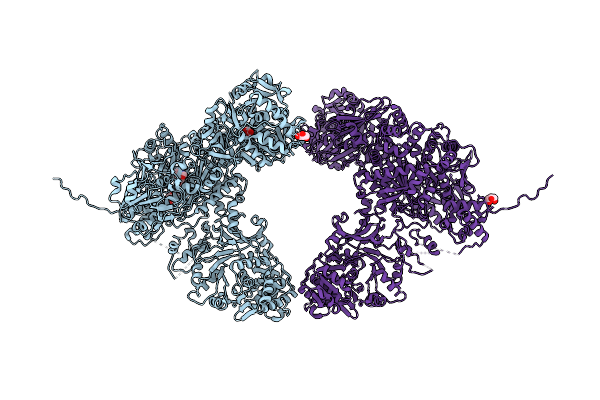 |
Organism: Brevibacillus parabrevis
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2024-11-20 Classification: BIOSYNTHETIC PROTEIN Ligands: GOL, A1AMN, MN |
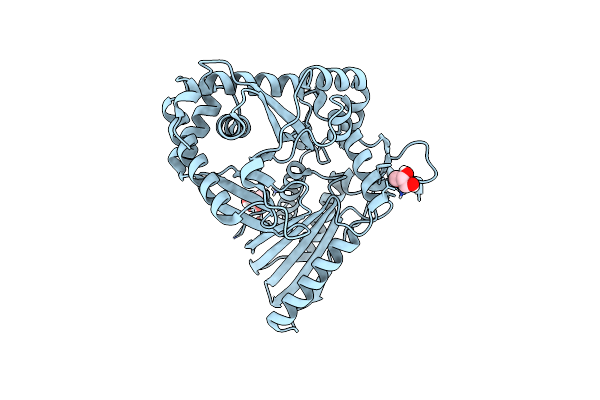 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2023-11-22 Classification: BIOSYNTHETIC PROTEIN Ligands: GOL |
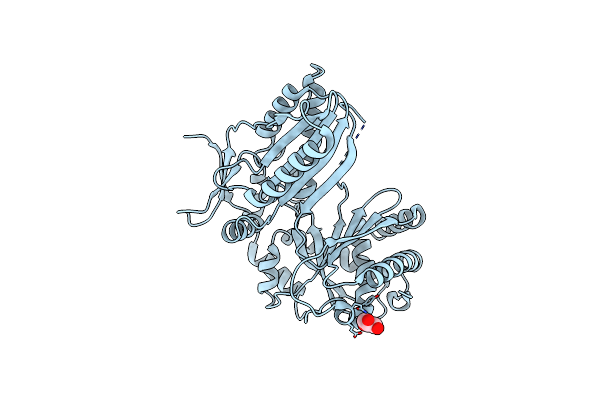 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2023-11-22 Classification: BIOSYNTHETIC PROTEIN Ligands: GOL |
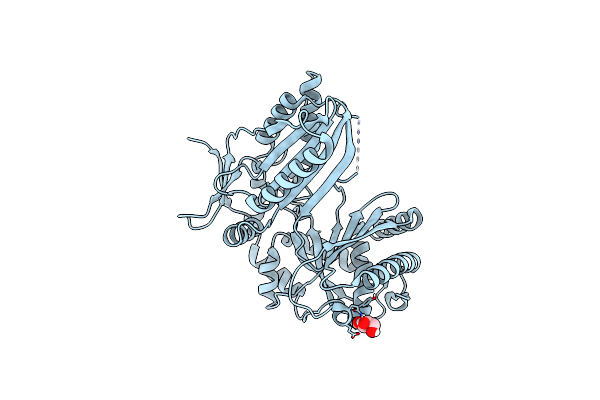 |
The Condensation Domain Of Surfactin A Synthetase C Variant 18B In Space Group P212121
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2023-11-22 Classification: BIOSYNTHETIC PROTEIN Ligands: GOL |
 |
The Condensation Domain Of Surfactin A Synthetase C Variant 18B In Space Group P43212
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2023-11-22 Classification: BIOSYNTHETIC PROTEIN Ligands: GOL |
 |
Organism: Stanieria sp. nies-3757
Method: ELECTRON MICROSCOPY Release Date: 2023-06-14 Classification: LIGASE |
 |
Cryo-Em Structure Of Stanieria Sp. Cpha2 In Complex With Adpcp And 4X(Beta-Asp-Arg)
Organism: stanieria sp. nies-3757, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-06-14 Classification: LIGASE Ligands: YHZ, MG, ACP |
 |
Crystal Structure Of Isoaspartyl Aminopeptidase From Roseivivax Halodurans Dsm 15395
Organism: Roseivivax halodurans
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2023-04-26 Classification: HYDROLASE |
 |
Organism: Leucothrix mucor dsm 2157
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-04-26 Classification: HYDROLASE Ligands: GOL, ZN, PO4 |
 |
Crystal Structure Of Wt Cyanophycin Dipeptide Hydrolase Cphz From Acinetobacter Baylyi Dsm587
Organism: Acinetobacter baylyi adp1
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2023-03-08 Classification: HYDROLASE Ligands: ZN, MN |
 |
Crystal Structure Of Cyanophycin Dipeptide Hydrolase Cphz E251A From Acinetobacter Baylyi Dsm587 In Complex With Beta-Asp-Arg
Organism: Acinetobacter baylyi adp1
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2023-03-08 Classification: HYDROLASE Ligands: 7ID, ZN, MN |

