Search Count: 28
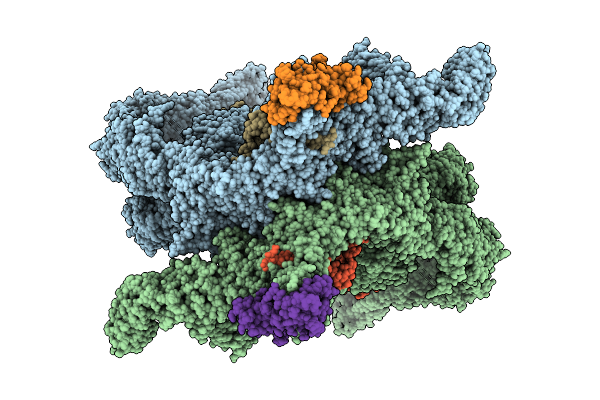 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: LIGASE Ligands: ZN |
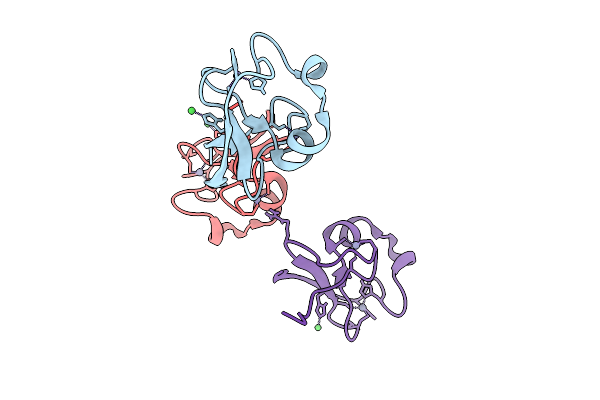 |
R-Degron Fused Zz-Domain Of The Arabidopsis Thaliana E3 Ubiquitin-Protein Ligase Big
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Release Date: 2025-09-17 Classification: LIGASE Ligands: ZN, NI |
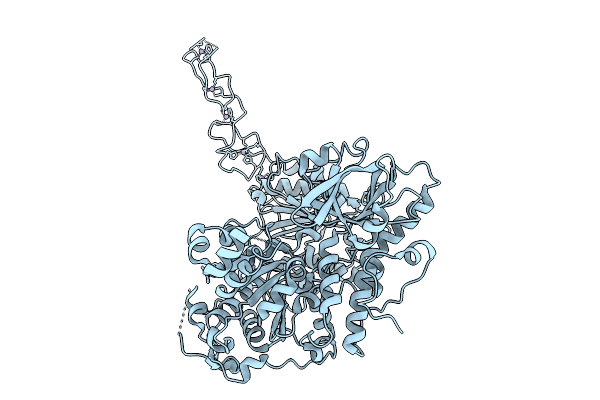 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: RNA BINDING PROTEIN Ligands: ZN |
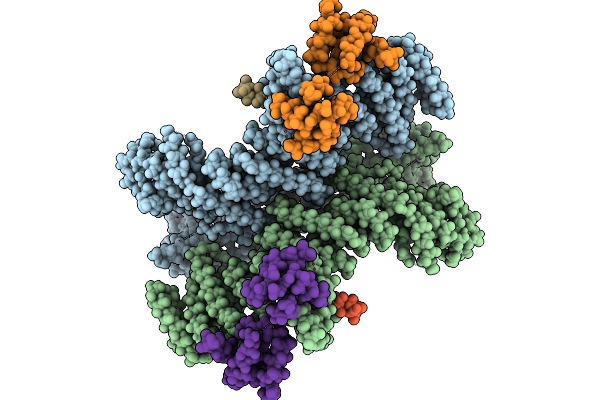 |
Cryo-Em Structure Of The Core Of The Arabidopsis Thaliana Ubr4/Di19/Calm1 Complex
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: LIGASE |
 |
Cryo-Em Structure Of The Human Ubr4/Kcmf1/Calm1 Complex (C-Term Dimer Interface Focused Refinement)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: LIGASE Ligands: ZN |
 |
Cryo-Em Structure Of The Human Ubr4/Kcmf1/Calm1 Complex (Calm1 Focused Refinement)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: LIGASE Ligands: ZN |
 |
Cryo-Em Structure Of The Human Ubr4/Kcmf1/Calm1 Complex (N-Term Focused Refinement)
|
 |
Cryo-Em Structure Of The Human Ubr4/Kcmf1/Calm1 Complex (Bp Focused Refinement)
|
 |
Cryo-Em Structure Of The Human Ubr4/Kcmf1/Calm1 Complex (C-Term Focused Refinement)
|
 |
Cryo-Em Structure Of The C. Elegans Ubr4/Kcmf1 Complex (C-Term Dimer Interface Focused Refinement)
Organism: Caenorhabditis elegans
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: LIGASE Ligands: ZN |
 |
Cryo-Em Structure Of The C. Elegans Ubr4/Kcmf1 Complex (N-Term Focused Refinement)
Organism: Caenorhabditis elegans
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: LIGASE |
 |
Cryo-Em Structure Of The C. Elegans Ubr4/Kcmf1 Complex (Side Focused Refinement)
Organism: Caenorhabditis elegans
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: LIGASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: PEPTIDE BINDING PROTEIN Ligands: ZN |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: LIGASE Ligands: ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-04-09 Classification: RNA BINDING PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-07-12 Classification: LIGASE Ligands: ZN |
 |
Organism: Spinacia oleracea
Method: ELECTRON MICROSCOPY Release Date: 2022-08-24 Classification: PLANT PROTEIN Ligands: ATP, ADP |
 |
Organism: Spinacia oleracea
Method: ELECTRON MICROSCOPY Release Date: 2022-05-25 Classification: PLANT PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.20 Å Release Date: 2021-09-01 Classification: TRANSPORT PROTEIN Ligands: K |
 |
Organism: Nematocida sp. ertm5
Method: X-RAY DIFFRACTION Resolution:3.04 Å Release Date: 2021-07-28 Classification: LIGASE |

