Search Count: 14
 |
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2025-02-19 Classification: STRUCTURAL PROTEIN Ligands: PO4 |
 |
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.31 Å Release Date: 2025-02-19 Classification: STRUCTURAL PROTEIN |
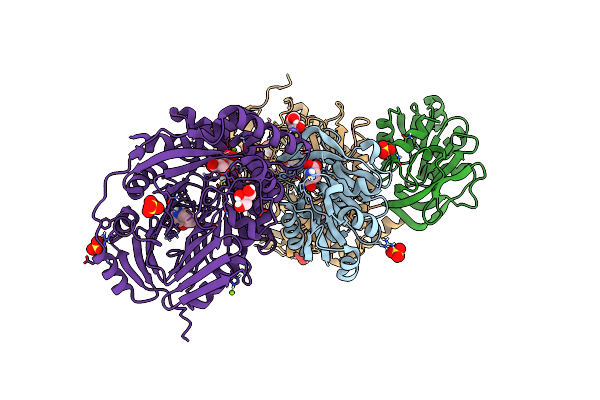 |
Aminodeoxychorismate Synthase Complex From Escherichia Coli, With Glutamine And Chorismate Added
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2025-01-29 Classification: TRANSFERASE Ligands: GLU, GOL, SO4, ISJ, TRP, MG, CL |
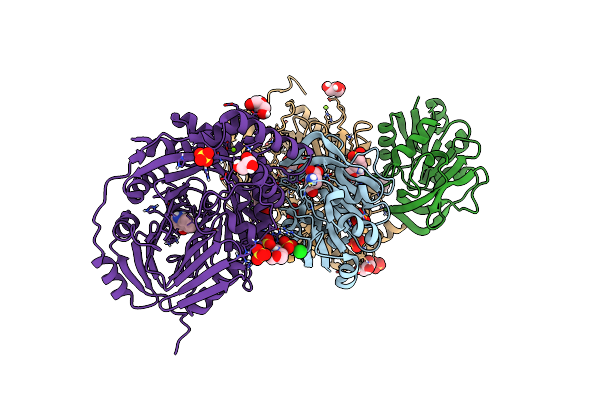 |
Aminodeoxychorismate Synthase Complex From Escherichia Coli, With Glutamine Added
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2025-01-29 Classification: TRANSFERASE Ligands: GLU, CL, SO4, MES, TRP, MG, GOL |
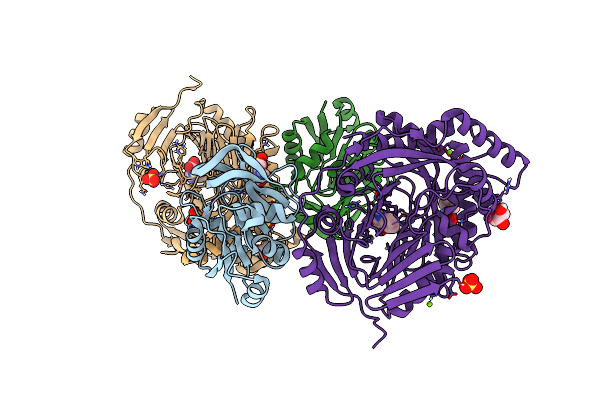 |
Aminodeoxychorismate Synthase Complex From Escherichia Coli, With Edta Added
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2025-01-29 Classification: TRANSFERASE Ligands: CL, SO4, TRP, MG, MES, GOL |
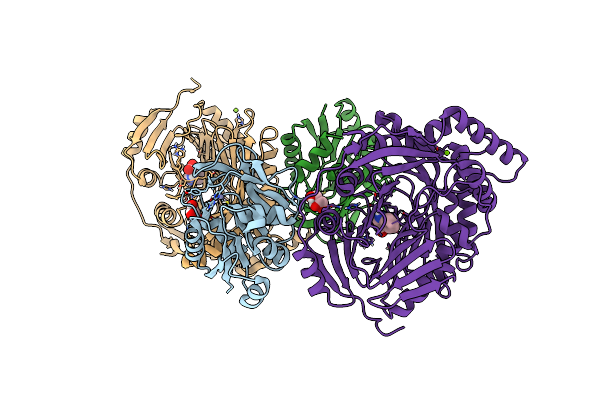 |
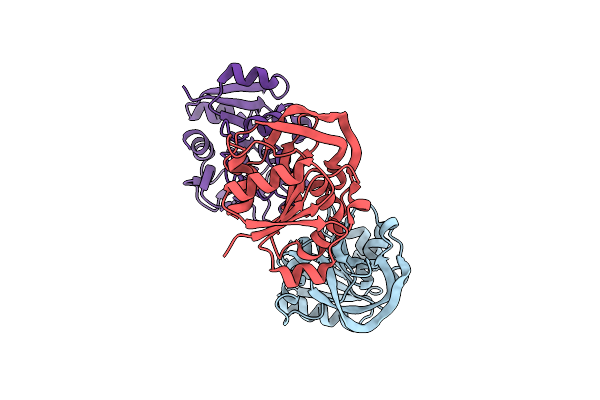
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-01-29 Classification: TRANSFERASE Ligands: ZN, GOL, TRP, MG, CL |
 |
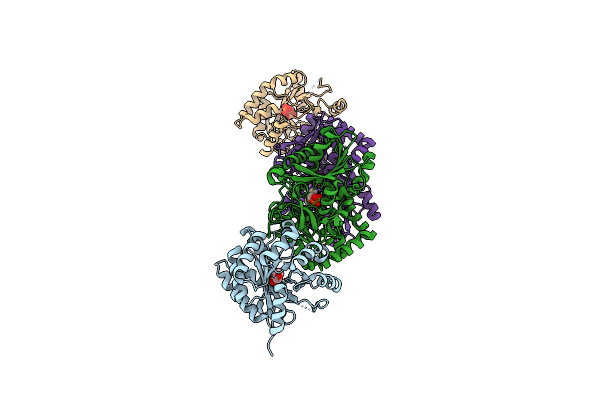
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.82 Å Release Date: 2025-01-29 Classification: TRANSFERASE |
 |
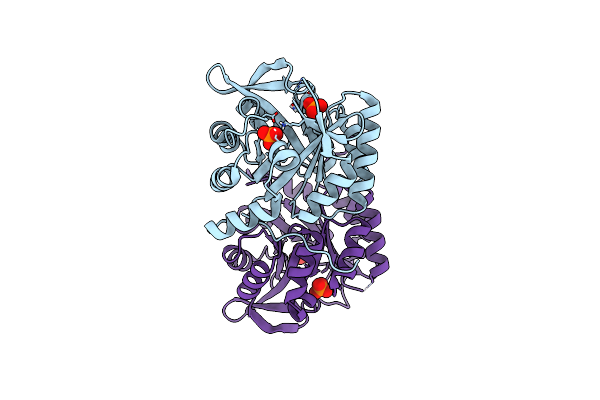
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2016-04-27 Classification: LYASE Ligands: G3P, PLP, NA |
 |
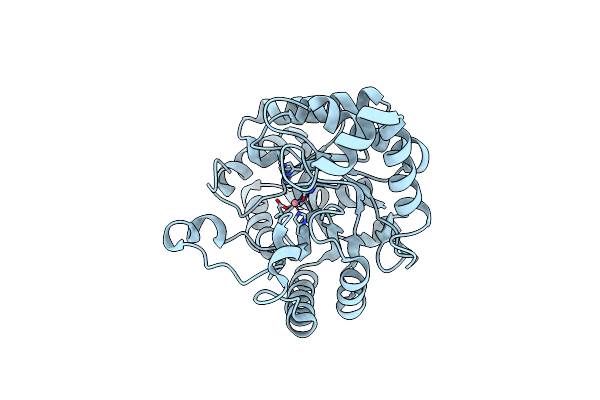
Organism: Artificial gene
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2013-11-06 Classification: LYASE Ligands: PI |
 |
Molecular Engineering Of Organophosphate Hydrolysis Activity From A Weak Promiscuous Lactonase Template
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2013-07-24 Classification: HYDROLASE Ligands: CO |
 |
Molecular Engineering Of Organophosphate Hydrolysis Activity From A Weak Promiscuous Lactonase Template
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2013-07-24 Classification: HYDROLASE Ligands: CO |
 |
Crystal Structure Of A Deinococcus Radiodurans Pte-Like Lactonase (Drpll) Mutant Y28L/D71N/E101G/E179D/V235L/L270M
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2013-07-24 Classification: HYDROLASE Ligands: CO |
 |
Anthranilate Phosphoribosyl-Transferase (Trpd) Double Mutant D83G F149S From S. Solfataricus
Organism: Sulfolobus solfataricus
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2009-12-15 Classification: TRANSFERASE Ligands: PRP, MN, PEG, GOL |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2006-01-31 Classification: CHAPERONE |

