Search Count: 148
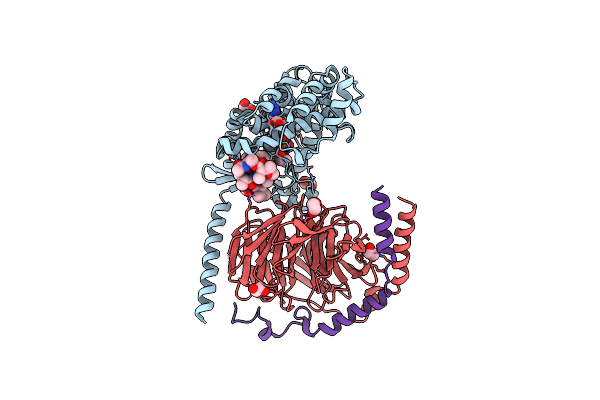 |
Crystal Structure Of The G11 Protein Heterotrimer Bound To Ym-254890 Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-03-19 Classification: SIGNALING PROTEIN Ligands: GDP, EDO, DAM, HF2, THC, OTH, HL2, MAA, ALA, ACE |
 |
Crystal Structure Of The G11 Protein Heterotrimer Bound To Fr900359 Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2025-03-19 Classification: SIGNALING PROTEIN Ligands: EDO, GDP, ZN, CL, DAM, HF2, UDL, OTH, HL2, MAA, ALA, PPI, DMS |
 |
Laser Excitation Effects On Br: Reprocessed Dark Dataset Recorded From Nango Et Al.
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-01-22 Classification: PROTON TRANSPORT Ligands: LI1, OLC, RET |
 |
Laser Excitation Effects On Br: Reprocessed Extrapolated 16 Ns Light Dataset Recorded At 0.04 Gw/Cm2 From Nango Et Al.
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-01-22 Classification: PROTON TRANSPORT Ligands: LI1, OLC, RET |
 |
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-01-22 Classification: PROTON TRANSPORT Ligands: LI1, OLC, RET |
 |
Laser Excitation Effects On Br: Extrapolated 6 Ps Light Dataset Recorded At 342 Gw/Cm2 At Swissfel
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-01-22 Classification: PROTON TRANSPORT Ligands: LI1, OLC, RET |
 |
Laser Excitation Effects On Br: Extrapolated 6 Ps Light Dataset Recorded At 2493 Gw/Cm2 At Swissfel
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-01-22 Classification: PROTON TRANSPORT Ligands: LI1, OLC, RET |
 |
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2025-01-22 Classification: PROTON TRANSPORT Ligands: LI1, OLC, RET |
 |
Laser Excitation Effects On Br: Reprocessed Extrapolated 10Ps Light Dataset Recorded At 525 Gw/Cm2 From Nogly Et Al.
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2025-01-22 Classification: PROTON TRANSPORT Ligands: LI1, OLC, RET |
 |
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2025-01-22 Classification: PROTON TRANSPORT Ligands: LI1, OLC, RET |
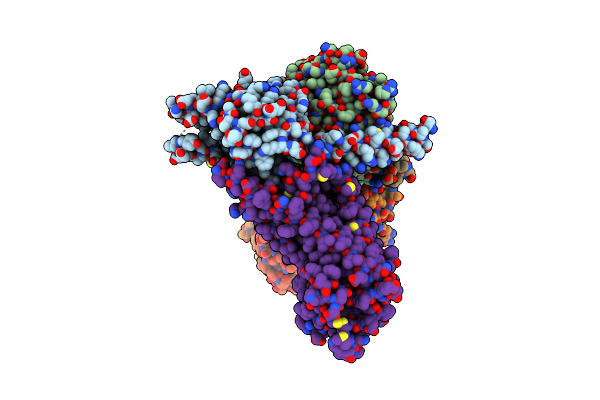
 |
Laser Excitation Effects On Br: Extrapolated 10Ps Light Dataset Recorded At 1281 Gw/Cm2 At Sacla (+100Ps, 1Ns, 10Ns)
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2025-01-22 Classification: PROTON TRANSPORT Ligands: LI1, OLC, RET |
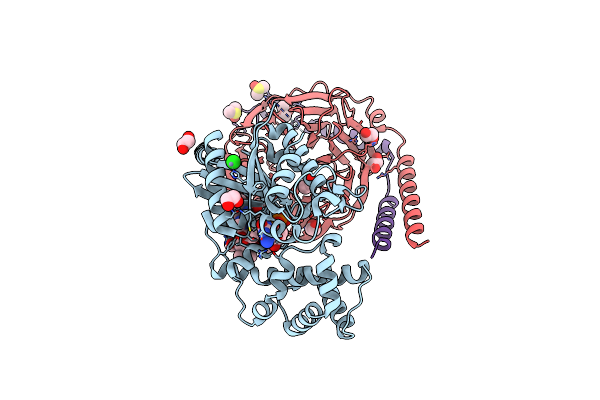
 |
Organism: Bos taurus, Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2024-11-20 Classification: SIGNALING PROTEIN |
 |
Cryo-Em Structure Of Rhodopsin-Gi Bound With Antibody Fragments Scfv16 And Fab79, Conformation 1
Organism: Bos taurus, Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2024-11-20 Classification: SIGNALING PROTEIN |
 |
Cryo-Em Structure Of Rhodopsin-Gi Bound With Antibody Fragments Scfv16 And Fab79, Conformation 2
Organism: Bos taurus, Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2024-11-20 Classification: SIGNALING PROTEIN |
 |
Organism: Homo sapiens, Hasarius adansoni, Bos taurus
Method: ELECTRON MICROSCOPY Release Date: 2024-10-30 Classification: MEMBRANE PROTEIN Ligands: RET |
 |
Cryo-Em Structure Of Jumping Spider Rhodopsin-1 Bound To A Giq Heterotrimer
Organism: Hasarius adansoni, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-10-23 Classification: MEMBRANE PROTEIN Ligands: A1H6M |
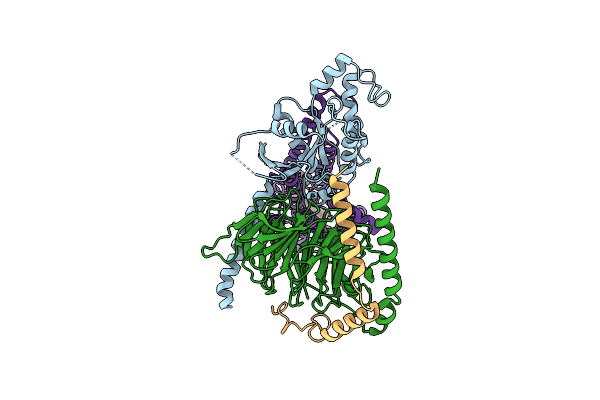 |
Cryo-Em Structure Of Jumping Spider Rhodopsin-1 Bound To A Giq Heterotrimer
Organism: Hasarius adansoni, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-10-23 Classification: MEMBRANE PROTEIN Ligands: A1H6M |
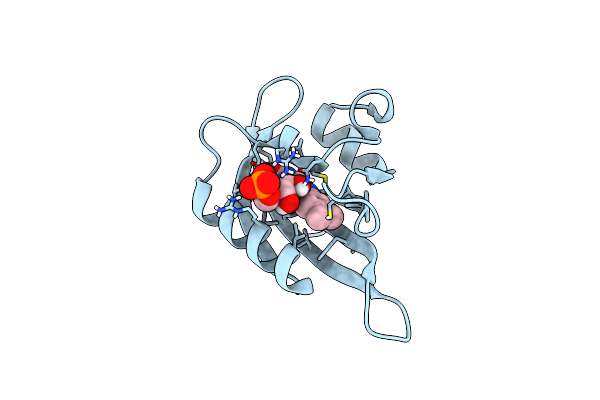 |
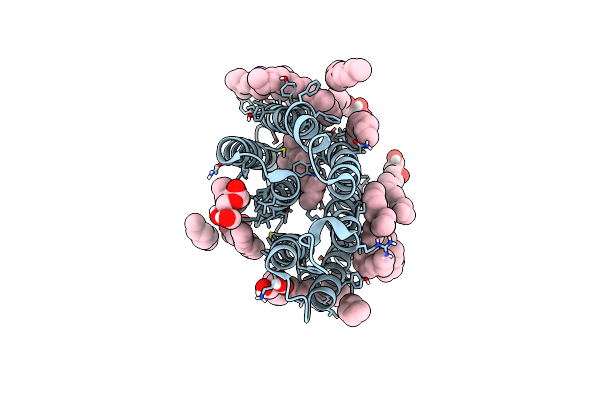
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2024-07-24 Classification: FLAVOPROTEIN Ligands: FMN |
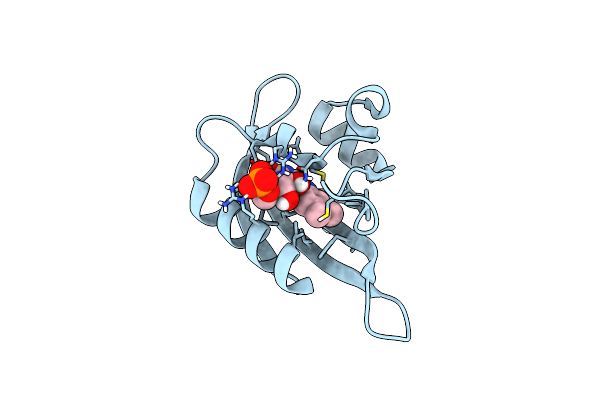 |
Crphotlov1 Dark State Structure Determined By Serial Synchrotron Crystallography At Room Temperature
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2024-07-24 Classification: FLAVOPROTEIN Ligands: FMN |
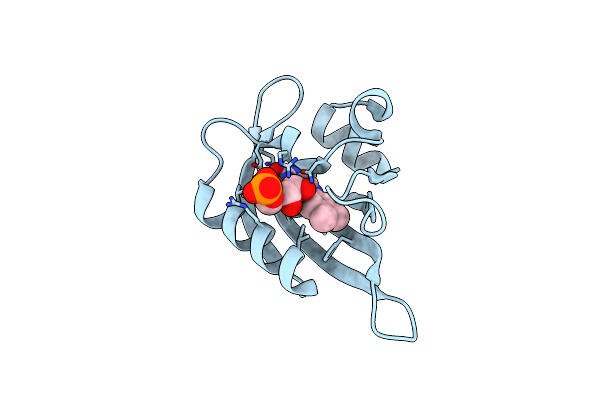 |
Crphotlov1 Light State Structure 2.5 Ms (0-5 Ms) After Illumination Determined By Time-Resolved Serial Synchrotron Crystallography At Room Temperature
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-07-24 Classification: FLAVOPROTEIN Ligands: FMN |

