Search Count: 85
 |
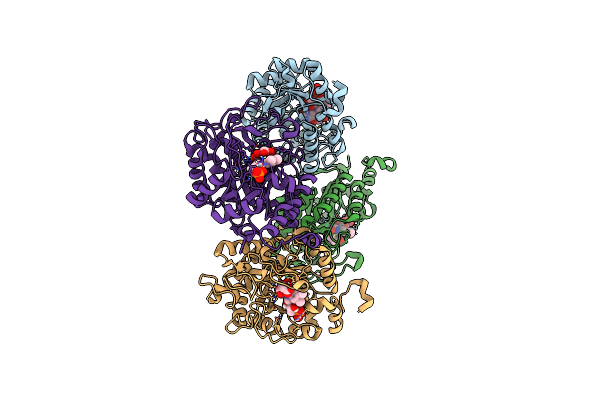
Ancestrally Reconstructed Acetolactate Synthase - Ancestor N259 With Bound Thdp And Magnesium.
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: TRANSFERASE Ligands: TPP, MG |
 |
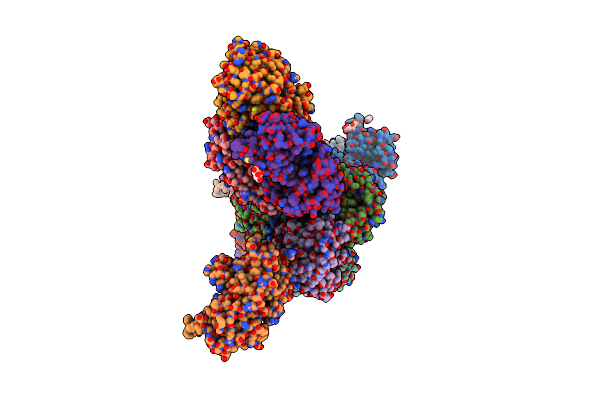
Crystal Structure Of Staphylococcus Aureus Ketol-Acid Reductoisomerase In Complex With Mg2+, And Jk-5-114
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Release Date: 2025-04-09 Classification: ISOMERASE Ligands: A1AKJ, MG |
 |
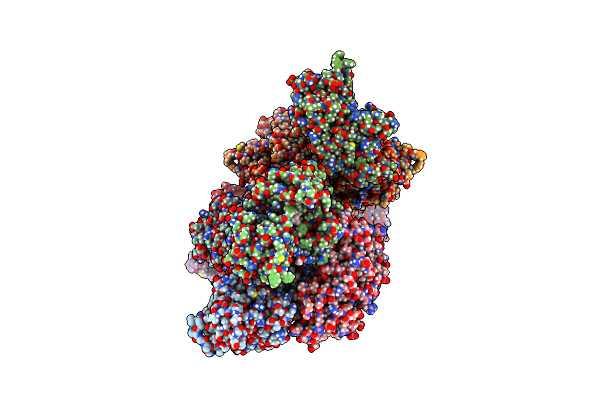
Crystal Structure Of Staphylococcus Aureus Ketol-Acid Reductoisomerase In Complex With Mg2+, And Jk-5-115
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Release Date: 2025-04-09 Classification: ISOMERASE Ligands: MG, A1AKY |
 |
Crystal Structure Of Oryza Sativa Ketol-Acid Reductoisomerase In Complex With Mg2+, And Jk-5-114
Organism: Oryza sativa japonica group
Method: X-RAY DIFFRACTION Release Date: 2025-04-09 Classification: ISOMERASE Ligands: MG, A1AKJ |
 |
Succinate Bound Crystal Structure Of Thermus Scotoductus Sa-01 Ene-Reductase
Organism: Thermus scotoductus sa-01
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2024-07-03 Classification: OXIDOREDUCTASE Ligands: SIN, FMN |
 |
Rhodococcus Ruber Alcohol Dehydrogenase Nadh Biomimetic Complex - Compound 4B
Organism: Rhodococcus ruber
Method: X-RAY DIFFRACTION Resolution:2.99 Å Release Date: 2024-07-03 Classification: OXIDOREDUCTASE Ligands: ZN, W46, CIT, IPA |
 |
Rhodococcus Ruber Alcohol Dehydrogenase Nadh Biomimetic Complex - Compound 1A
Organism: Rhodococcus ruber
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-07-03 Classification: OXIDOREDUCTASE Ligands: IPA, ZN, NA, W3O, CIT |
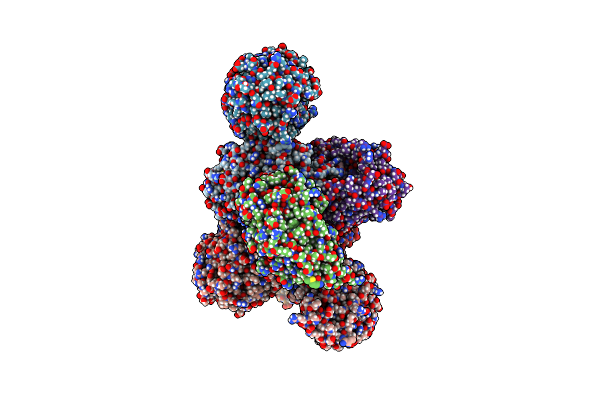 |
Organism: Thermus scotoductus sa-01
Method: X-RAY DIFFRACTION Resolution:2.76 Å Release Date: 2024-07-03 Classification: FLAVOPROTEIN Ligands: FMN, W3X, IPA, NA, CL |
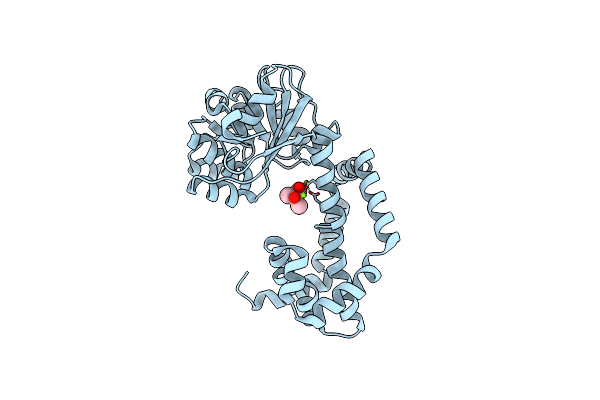 |
Crystal Structure Of Campylobacter Jejuni Ketol-Acid Reductoisomerase In Complex With 2-Acetolactate
Organism: Campylobacter jejuni subsp. jejuni
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2024-04-24 Classification: OXIDOREDUCTASE Ligands: MG, X2X, CL |
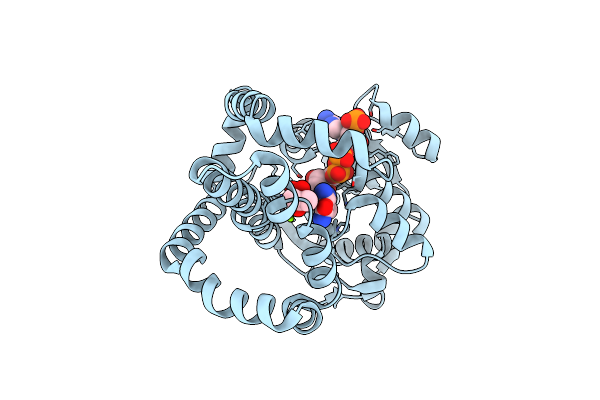 |
Campylobacter Jejuni Keto-Acid Reductoisomerase In Complex With Intermediate And Nadp+
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2024-04-24 Classification: OXIDOREDUCTASE Ligands: WXU, MG, NCA, NDP, CL |
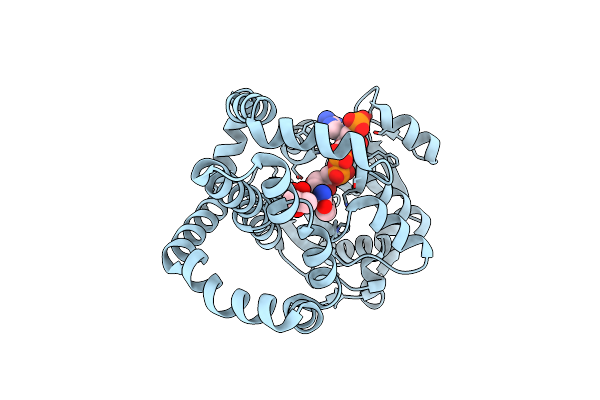 |
Campylobacter Jejuni Ketol-Acid Reductoisomerase In Complex With Nadp+ And Hmkb
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2024-04-24 Classification: OXIDOREDUCTASE Ligands: MG, CL, NDP, WXU |
 |
Campylobacter Jejuni Ketol-Acid Reductoisomerase In Complex With Nadph And Hoe704
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2024-04-24 Classification: OXIDOREDUCTASE Ligands: MG, NDP, X9W |
 |
Campylobacter Jejuni Ketol-Acid Reductoisomerase In Complex With 2,3-Dihydroxy-3-Isovalerate.
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2024-04-24 Classification: OXIDOREDUCTASE Ligands: MG, XAI |
 |
Crystal Structure Of Fe-S Cluster-Dependent Dehydratase From Paralcaligenes Ureilyticus In Complex With Mg
Organism: Paralcaligenes ureilyticus
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2023-08-30 Classification: LYASE Ligands: FES, MG, BCT, CO2 |
 |
Crystal Structure Of Red Kidney Bean Purple Acid Phosphatase In Complex With An Alpha-Aminonaphthylmethylphosphonic Acid Inhibitor
Organism: Phaseolus vulgaris
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-08-02 Classification: METAL BINDING PROTEIN Ligands: ZN, FE, FLC, PGE, NAG, SO4, EDO, GOL, CL, R9X, NA, DMS |
 |
Crystal Structure Of Wild-Type Arabidopsis Thaliana Acetohydroxyacid Synthase In Complex With Amidosulfuron
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2023-03-29 Classification: TRANSFERASE Ligands: MG, FAD, WRQ, TLA, NHE, AUJ, SO4 |
 |
Crystal Structure Of Arabidopsis Thaliana Acetohydroxyacid Synthase S653T Mutant In Complex With Amidosulfuron
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.94 Å Release Date: 2023-03-29 Classification: TRANSFERASE Ligands: MG, FAD, WRQ, NHE, AUJ, TLA, SO4 |
 |
Apo Form Cryo-Em Structure Of Campylobacter Jejune Ketol-Acid Reductoisommerase Crosslinked By Glutaraldehyde
Organism: Campylobacter jejuni
Method: ELECTRON MICROSCOPY Release Date: 2023-02-01 Classification: ISOMERASE Ligands: PTD |
 |
Organism: Synechococcus elongatus pcc 7942 = fachb-805
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2022-08-24 Classification: TRANSFERASE Ligands: IPE, EDO, PEG, SO4, MG, CL |
 |
Organism: Synechocystis sp. (strain pcc 6803 / kazusa)
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2022-06-01 Classification: TRANSFERASE Ligands: MG, CL |

