Search Count: 42
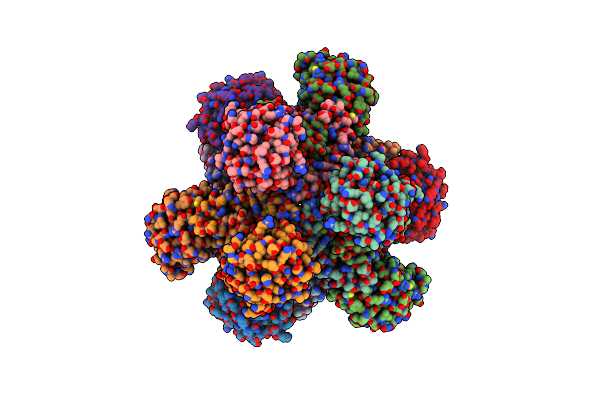 |
Apo Form Cryo-Em Structure Of Campylobacter Jejune Ketol-Acid Reductoisommerase Crosslinked By Glutaraldehyde
Organism: Campylobacter jejuni
Method: ELECTRON MICROSCOPY Release Date: 2023-02-01 Classification: ISOMERASE Ligands: PTD |
 |
The Crystal Structure Of The Tir Domain-Containing Protein From Acinetobacter Baumannii (Abtir)
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2022-09-07 Classification: HYDROLASE Ligands: SO4, P6G |
 |
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2022-09-07 Classification: HYDROLASE |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2022-09-07 Classification: HYDROLASE Ligands: OJC, GOL, SO4 |
 |
Organism: Streptococcus equi
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2022-09-07 Classification: HYDROLASE Ligands: PGE, GOL |
 |
Organism: Acinetobacter baumannii
Method: ELECTRON MICROSCOPY Release Date: 2022-09-07 Classification: HYDROLASE Ligands: 1O4 |
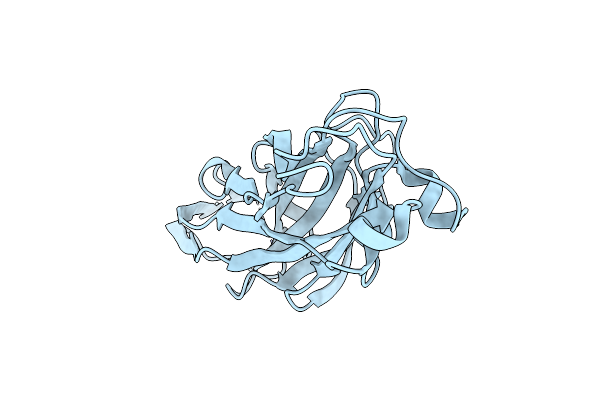 |
Organism: Proteus mirabilis
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2022-07-06 Classification: SUGAR BINDING PROTEIN Ligands: CL |
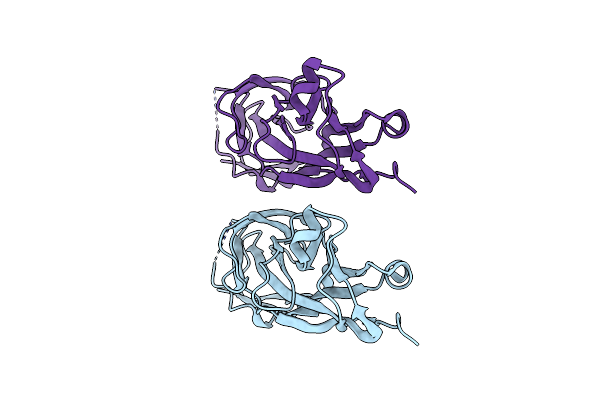 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-07-06 Classification: SUGAR BINDING PROTEIN Ligands: IOD |
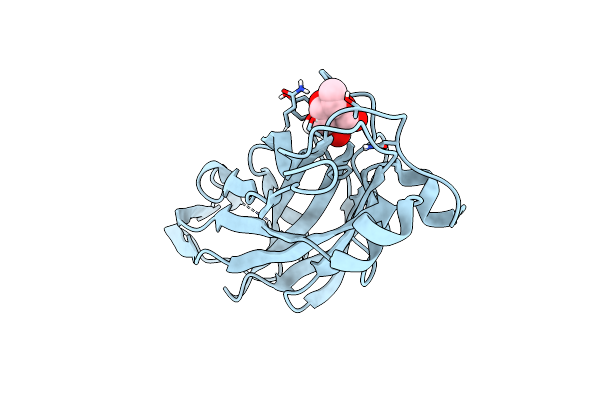 |
Organism: Proteus mirabilis
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-07-06 Classification: SUGAR BINDING PROTEIN Ligands: FUL, CL |
 |
Crystal Structure Of The Ucad Lectin-Binding Domain In Complex With Glucose
Organism: Proteus mirabilis
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2022-07-06 Classification: SUGAR BINDING PROTEIN Ligands: BGC, CL |
 |
Crystal Structure Of The Ucad Lectin-Binding Domain In Complex With Galactose
Organism: Proteus mirabilis
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2022-07-06 Classification: SUGAR BINDING PROTEIN Ligands: GLA, CL |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2022-03-09 Classification: MICROBIAL PROTEIN Ligands: GOL, IPA, CIT, PEG |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2022-03-09 Classification: MICROBIAL PROTEIN Ligands: GOL |
 |
Organism: Escherichia coli o157:h7
Method: X-RAY DIFFRACTION Resolution:2.98 Å Release Date: 2022-03-09 Classification: MICROBIAL PROTEIN Ligands: PEG, P6G, GOL, MG, CL, SO4 |
 |
Organism: Pontibacter korlensis
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2021-06-23 Classification: HYDROLASE Ligands: EPE, ZN, NA, SO4 |
 |
Crystal Structure Of Selenomethionine-Containing Acab From Uropathogenic E. Coli
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2020-07-15 Classification: TRANSCRIPTION Ligands: CL |
 |
Crystal Structure Of Transcription Regulator Acab From Uropathogenic E. Coli
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.94 Å Release Date: 2020-07-15 Classification: TRANSCRIPTION Ligands: CA |
 |
Organism: Escherichia coli o6:h1
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2019-04-10 Classification: PROTEIN BINDING Ligands: GOL, P6G, SO4, CA, CL, NA |
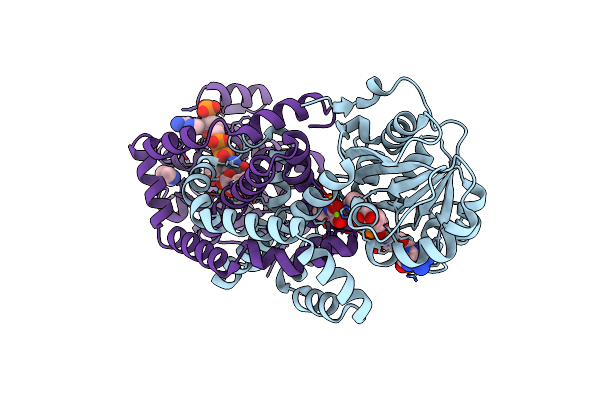 |
Crystal Structure Of Staphylococcus Aureus Ketol-Acid Reductoisomerase With Hydroxyoxamate Inhibitor 1
Organism: Staphylococcus aureus (strain bovine rf122 / et3-1)
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2019-01-30 Classification: OXIDOREDUCTASE Ligands: NDP, MG, EN4, 81B, IMD |
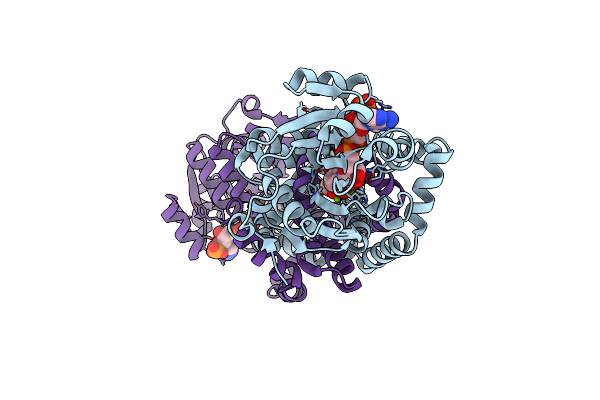 |
Crystal Structure Of Staphylococcus Aureus Ketol-Acid Reductoisomerase With Hydroxyoxamate Inhibitor 2
Organism: Staphylococcus aureus (strain bovine rf122 / et3-1)
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2018-12-12 Classification: Oxidoreductase/Inhibitor Ligands: MG, NAP, E9G, E9J |

