Search Count: 11
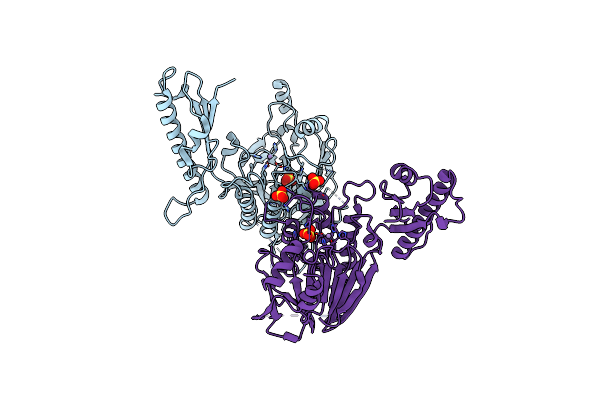 |
Crystal Structure Of A Covalently Fused Nbs1-Mre11 Complex With One Manganese Ion Per Active Site
Organism: Schizosaccharomyces pombe
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2012-06-20 Classification: HYDROLASE, PROTEIN BINDING Ligands: SO4, MN |
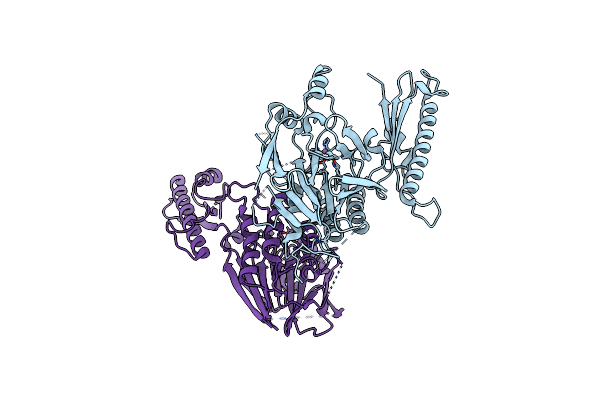 |
Crystal Structure Of A Covalently Fused Nbs1-Mre11 Complex With Two Manganese Ions Per Active Site
Organism: Schizosaccharomyces pombe
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2012-06-20 Classification: HYDROLASE, PROTEIN BINDING Ligands: MN |
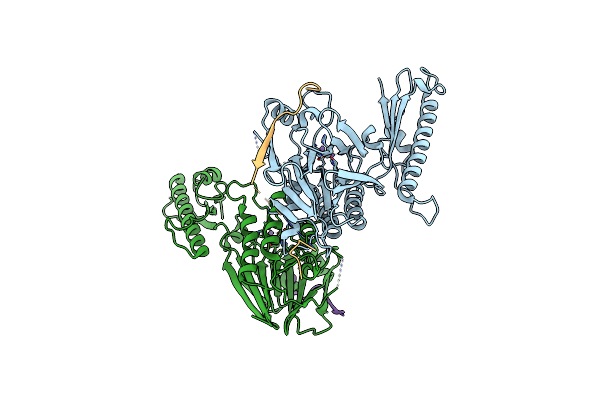 |
Crystal Structure Of An Unfused Mre11-Nbs1 Complex With Two Manganese Ions Per Active Site
Organism: Schizosaccharomyces pombe
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2012-06-20 Classification: HYDROLASE/PROTEIN BINDING Ligands: MN |
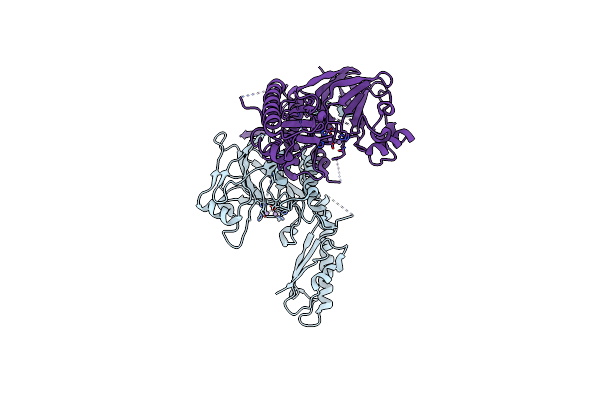 |
Organism: Schizosaccharomyces pombe
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2012-06-20 Classification: HYDROLASE Ligands: MN |
 |
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.81 Å Release Date: 2011-10-12 Classification: HYDROLASE Ligands: MN, SO4 |
 |
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2011-10-12 Classification: HYDROLASE/DNA BINDING PROTEIN Ligands: ADP, MG, SO4, PO4, MN |
 |
The Mre11:Rad50 Complex Forms An Atp Dependent Molecular Clamp In Dna Double-Strand Break Repair
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2011-04-20 Classification: HYDROLASE Ligands: ANP, MG |
 |
The Mre11:Rad50 Complex Forms An Atp Dependent Molecular Clamp In Dna Double-Strand Break Repair
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2011-04-20 Classification: HYDROLASE Ligands: GOL |
 |
Structure Of The Complete Abce1/Rnaase-L Inhibitor Protein From Pyrococcus Abysii
Organism: Pyrococcus abyssi
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2007-12-25 Classification: HYDROLYASE/TRANSLATION Ligands: MG, SF4, ADP |
 |
Structural Biochemistry Of Atp-Driven Dimerization And Dna Stimulated Activation Of Smc Atpases.
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2004-12-07 Classification: CELL CYCLE |
 |
Structural Biochemistry Of Atp-Driven Dimerization And Dna Stimulated Activation Of Smc Atpases.
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2004-12-07 Classification: CELL CYCLE Ligands: MG, ATP |

