Search Count: 107
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In Its Pfr State (I0A).
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, CL, EDO |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In Its Pfr State (I0B).
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4 |
 |
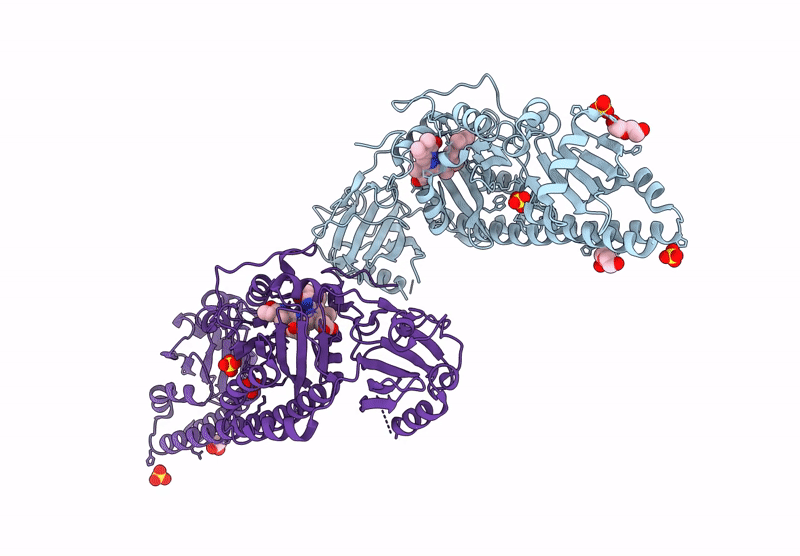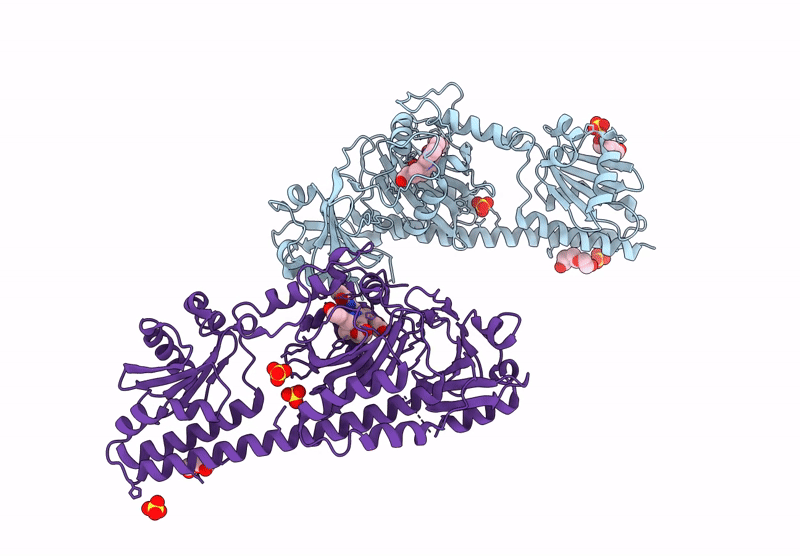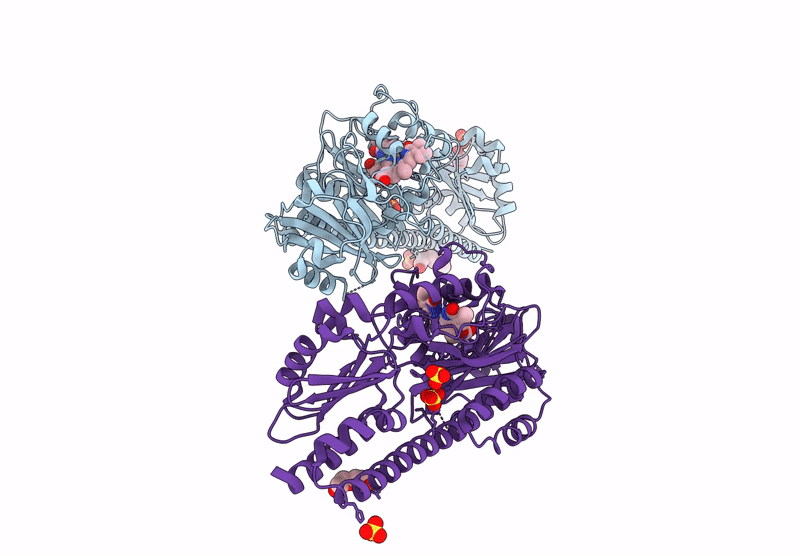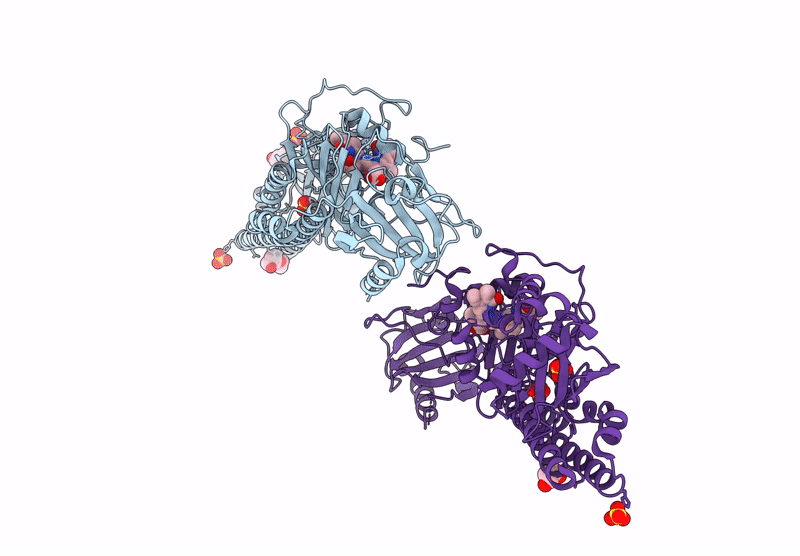
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I1 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4 |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I2 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, PGE, PEG, CL |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I3 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, GOL, PEG |
 |
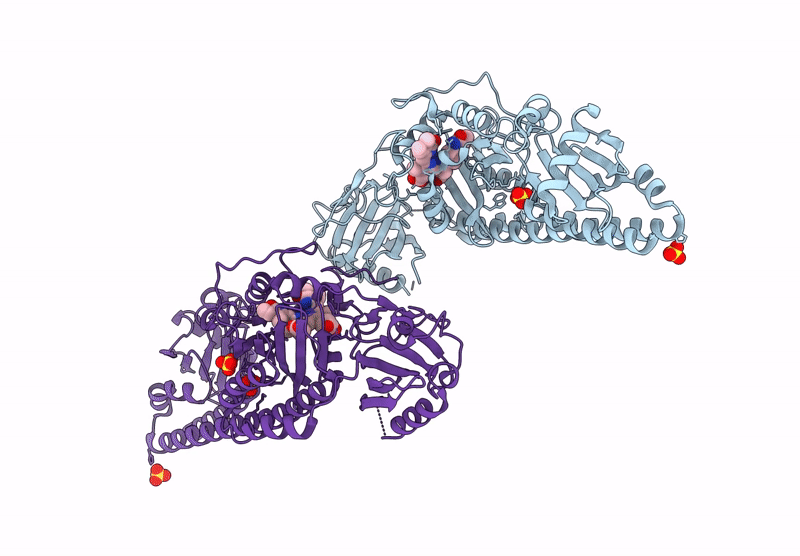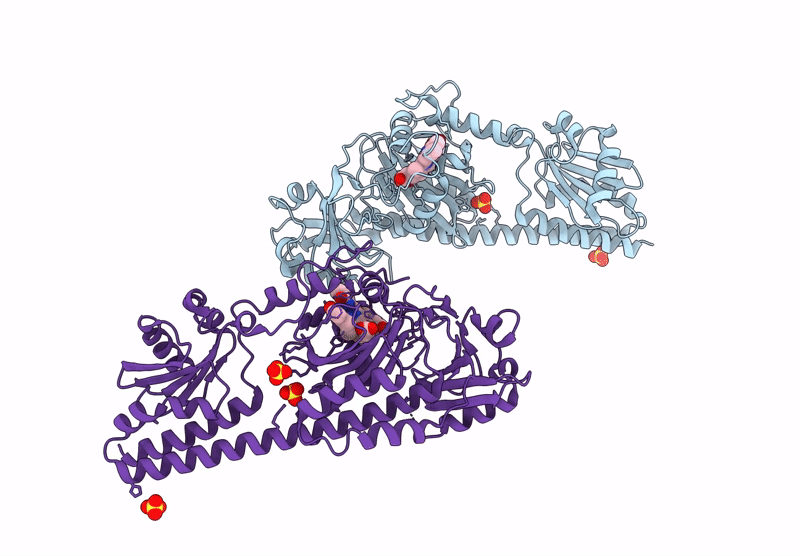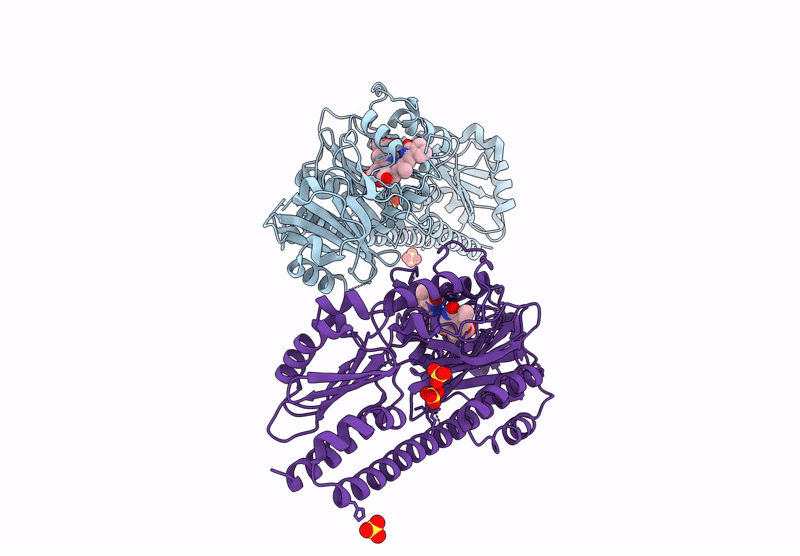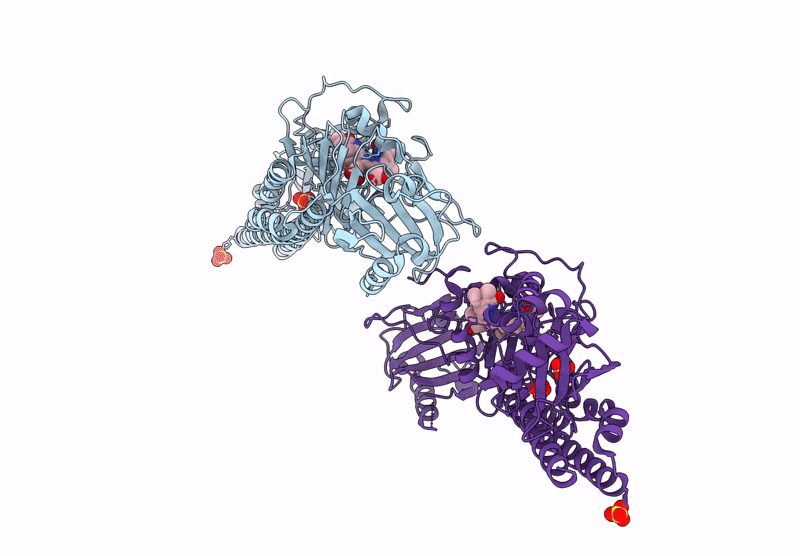
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I4 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, CL, PEG |
 |
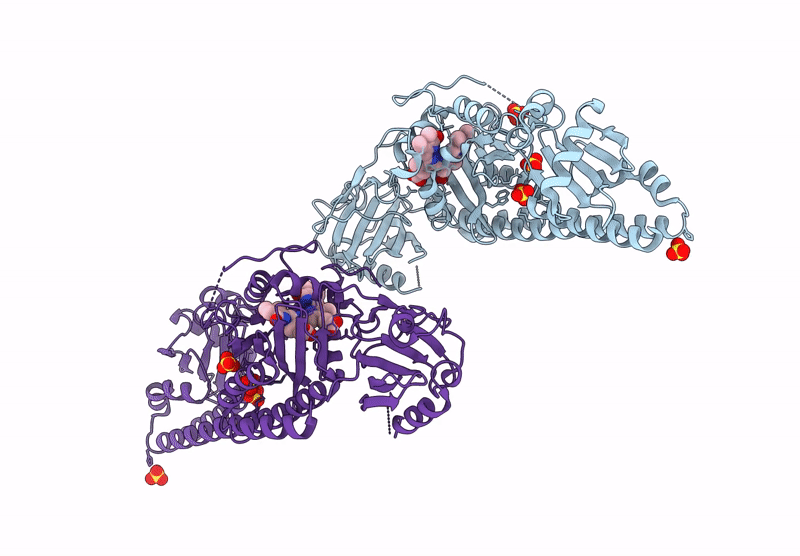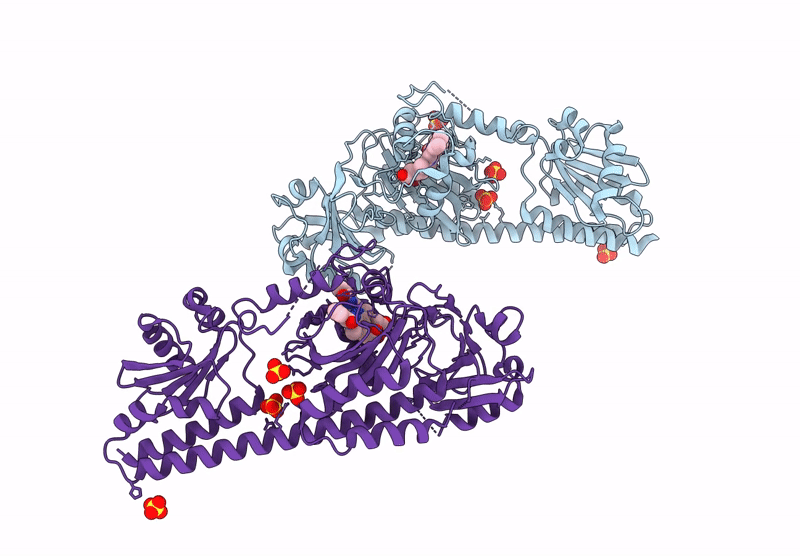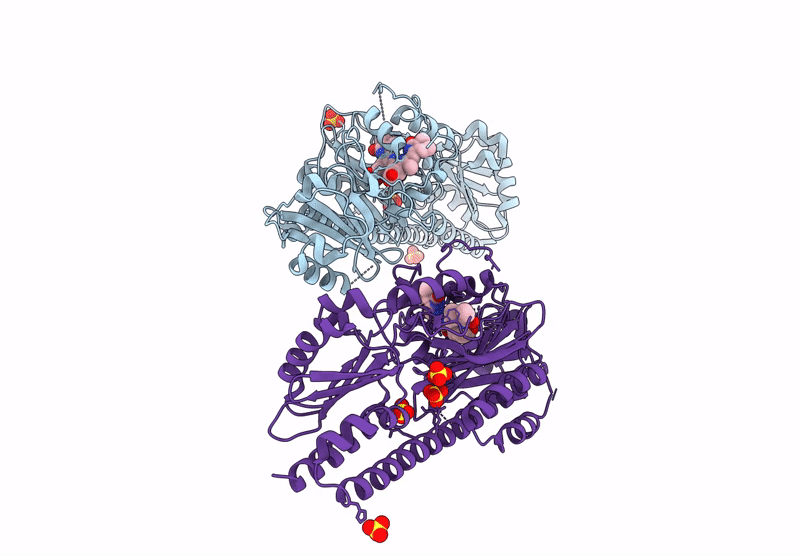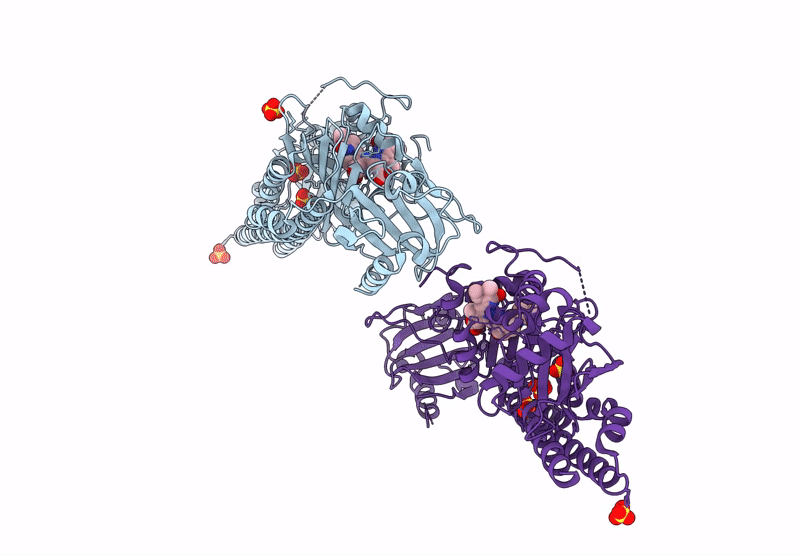
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I5 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, CL |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I6 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, CL |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I7 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, PEG |
 |
Crystal Structure Of The Light-Driven Proton Pump Heimdallarchaeial Rhodopsin Heimdallr1
Organism: Candidatus heimdallarchaeota archaeon
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-04-02 Classification: PROTON TRANSPORT Ligands: RET, OLC, NO3 |
 |
Organism: Synechocystis
Method: X-RAY DIFFRACTION Resolution:3.70 Å Release Date: 2025-03-12 Classification: FLUORESCENT PROTEIN Ligands: CYC |
 |
Kalium Channelrhodopsin 1 C110A Mutant From Hyphochytrium Catenoides, Dark State
Organism: Hyphochytrium catenoides
Method: ELECTRON MICROSCOPY Release Date: 2025-02-19 Classification: MEMBRANE PROTEIN Ligands: RET, CLR, PEE |
 |
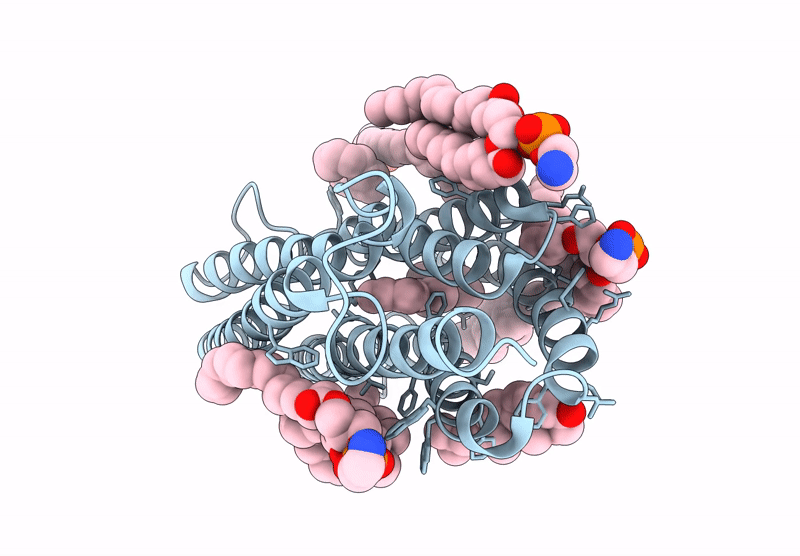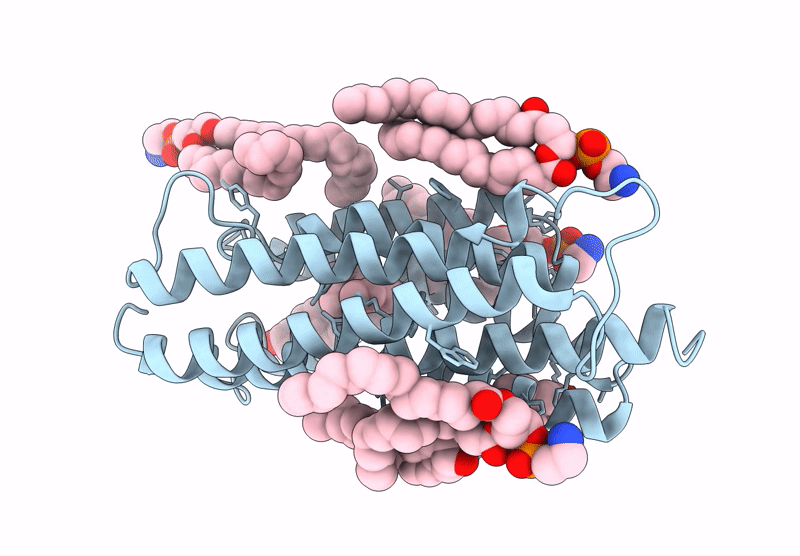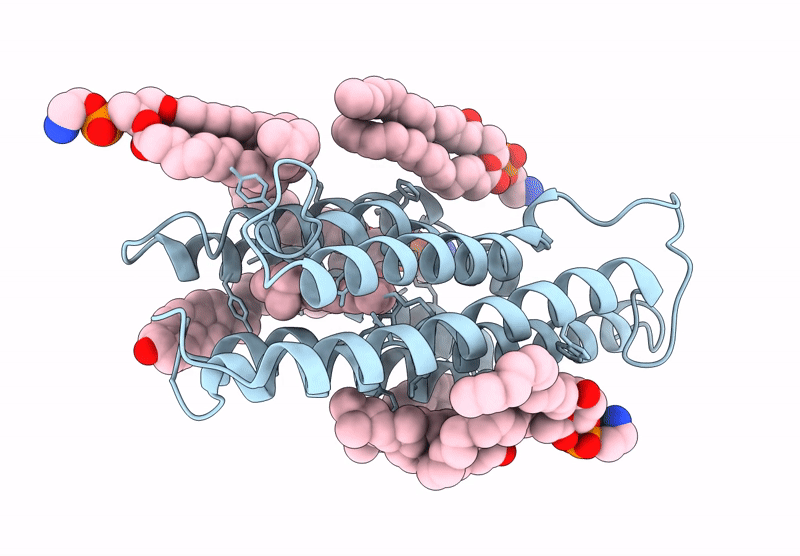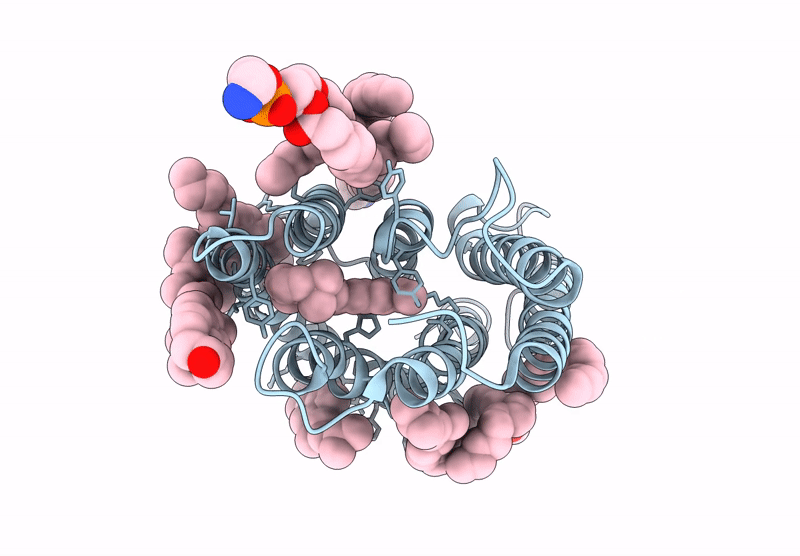
Kalium Channelrhodopsin 1 C110A Mutant From Hyphochytrium Catenoides, Laser-Flash-Illuminated
Organism: Hyphochytrium catenoides
Method: ELECTRON MICROSCOPY Release Date: 2025-02-19 Classification: MEMBRANE PROTEIN Ligands: RET, CLR, PEE |
 |
Kalium Channelrhodopsin 1 C110A Mutant From Hyphochytrium Catenoides, Continuous Illumination State
Organism: Hyphochytrium catenoides
Method: ELECTRON MICROSCOPY Release Date: 2025-02-19 Classification: MEMBRANE PROTEIN Ligands: RET, CLR, PEE |
 |
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2025-01-29 Classification: MEMBRANE PROTEIN Ligands: OLC, RET |
 |
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2025-01-29 Classification: MEMBRANE PROTEIN Ligands: OLC, RET |
 |
Ultrafast Structural Transitions In An Azobenzene Photoswitch At Near-Atomic Resolution: 1 Ns Structure
Organism: Synthetic construct, Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-11-13 Classification: CELL CYCLE Ligands: GTP, MG, CA, GDP, IBL |
 |
Ultrafast Structural Transitions In An Azobenzene Photoswitch At Near-Atomic Resolution: 10 Ns Structure
Organism: Synthetic construct, Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-11-13 Classification: CELL CYCLE Ligands: GTP, MG, CA, GDP, IBL |
 |
Ultrafast Structural Transitions In An Azobenzene Photoswitch At Near-Atomic Resolution: Dark Structure
Organism: Synthetic construct, Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-10-30 Classification: CELL CYCLE Ligands: MG, GTP, CA, IBL |
 |
Ultrafast Structural Transitions In An Azobenzene Photoswitch At Near-Atomic Resolution: 233 Fs Structure
Organism: Synthetic construct, Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-10-02 Classification: CELL CYCLE Ligands: GTP, MG, CA, IBL |

