Search Count: 13
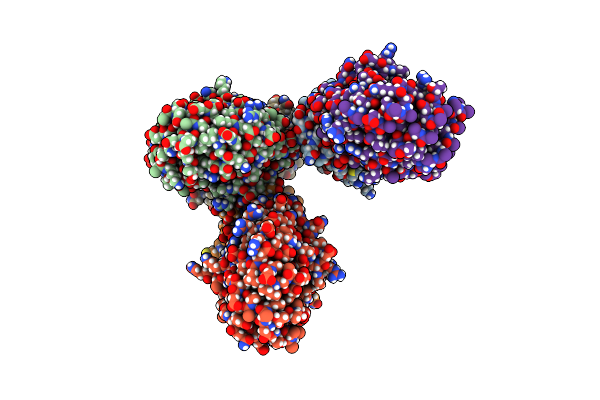 |
Crystal Structure Of Archaeoglobus Fulgidus (S)-3-O-Geranylgeranylglyceryl Phosphate Synthase
Organism: Archaeoglobus fulgidus dsm 4304
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2024-10-16 Classification: TRANSFERASE Ligands: PO4, SO4 |
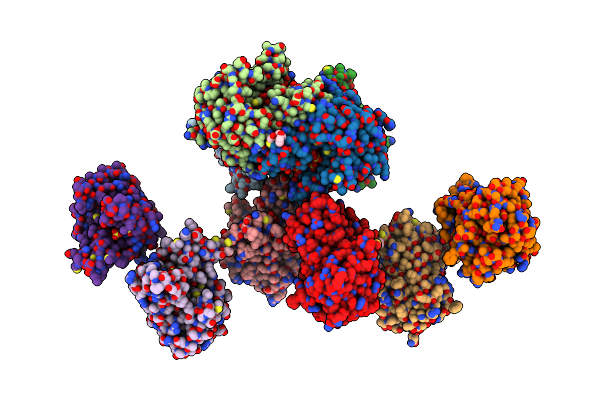 |
Crystal Structure Of The Halohydrin Dehalogenase Hheg D114C Mutant Cross-Linked With Bmoe
Organism: Ilumatobacter coccineus ym16-304
Method: X-RAY DIFFRACTION Release Date: 2022-03-09 Classification: LYASE Ligands: ME7, SO4 |
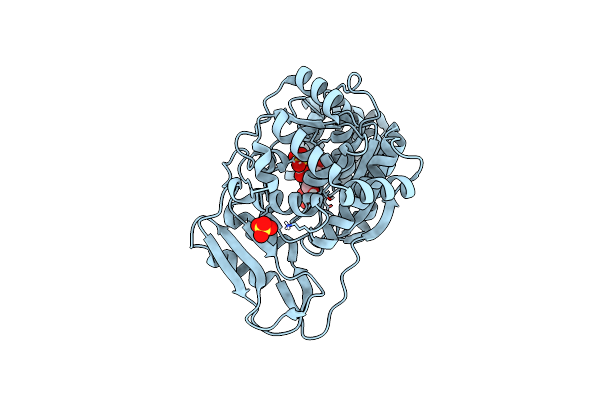 |
Crystal Structure Of Uridine Bound To Geobacillus Thermoglucosidasius Pyrimidine Nucleoside Phosphorylase Pynp
Organism: Parageobacillus thermoglucosidasius
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2021-08-25 Classification: TRANSFERASE Ligands: SO4, URI |
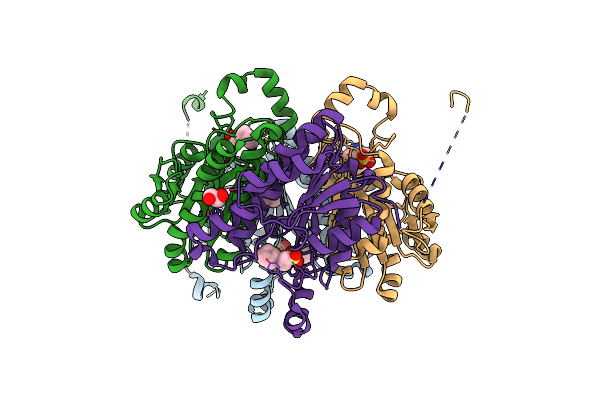 |
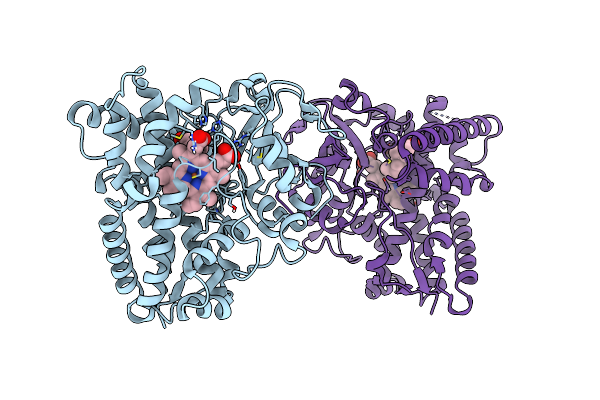
Insight Into The Molecular Determinants Of Thermal Stability In Halohydrin Dehalogenase Hhed2.
Organism: Gamma proteobacterium htcc2207
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2021-04-07 Classification: LYASE Ligands: NHE, GOL |
 |
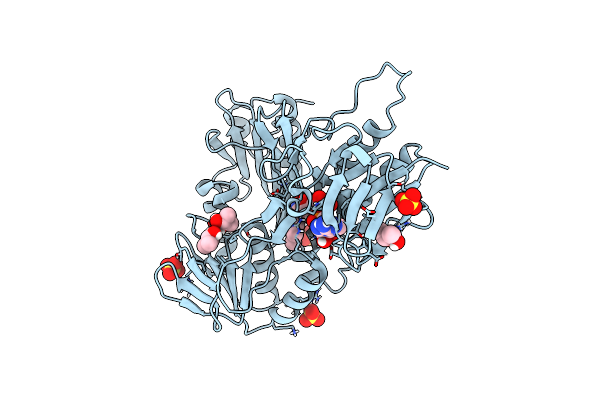
Crystal Structure Of Cyp154C5 From Nocardia Farcinica In Complex With 5Alpha-Androstan-3-One
Organism: Nocardia farcinica ifm 10152
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-11-18 Classification: OXIDOREDUCTASE Ligands: HEM, NQ8, CL |
 |
Organism: Thermobifida fusca yx
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-11-18 Classification: OXIDOREDUCTASE Ligands: FAD, BU3, SO4 |
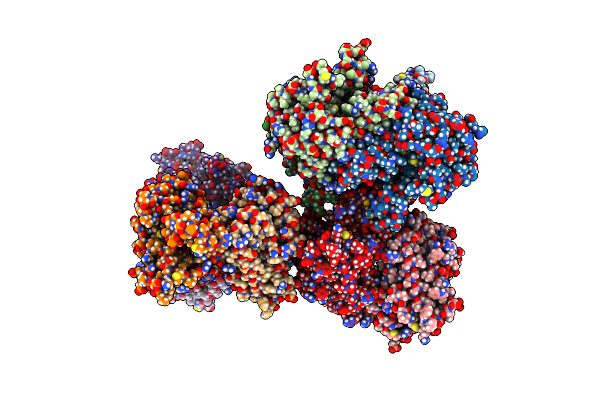 |
Organism: Ilumatobacter coccineus ym16-304
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2019-08-21 Classification: LYASE |
 |
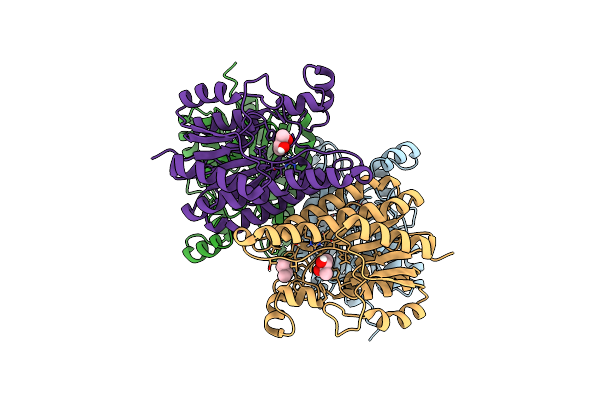
Organism: Ilumatobacter coccineus ym16-304
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2019-08-21 Classification: LYASE |
 |
Organism: Ilumatobacter coccineus ym16-304
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2019-08-21 Classification: LYASE Ligands: BU3, CL |
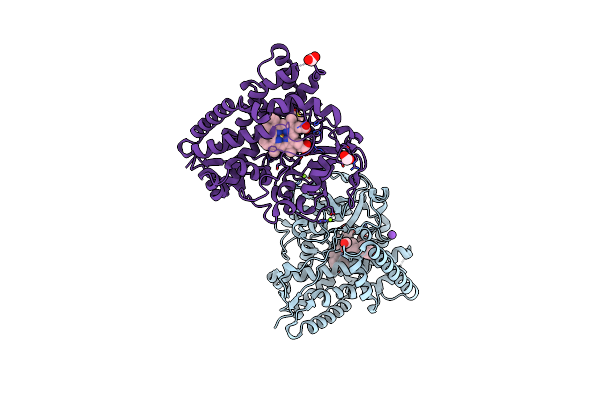 |
The 2.2 A Crystal Structure Of Cyp154C5 From Nocardia Farcinica In Complex With Pregnenolone
Organism: Nocardia farcinica
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2014-03-05 Classification: oxidoreductase/substrate Ligands: PLO, HEM, MG, FMT, K |
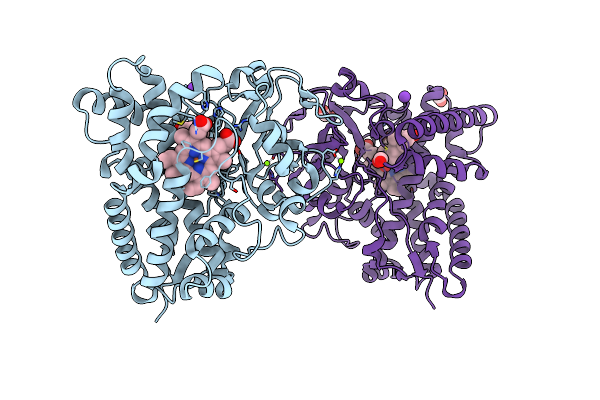 |
The 1.9 A Crystal Structure Of Cyp154C5 From Nocardia Farcinica In Complex With Progesterone
Organism: Nocardia farcinica
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2014-03-05 Classification: oxidoreductase/substrate Ligands: STR, HEM, MG, FMT, K |
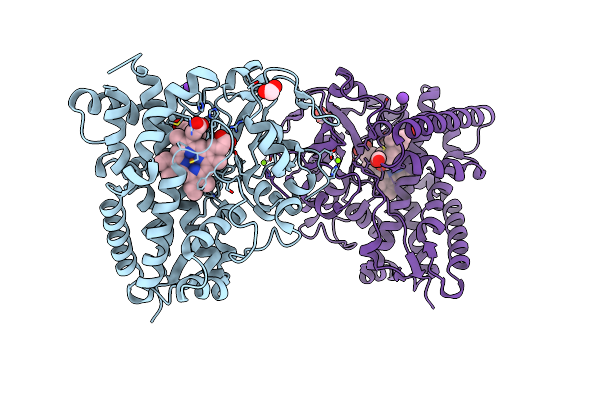 |
The 2.4 A Crystal Structure Of Cyp154C5 From Nocardia Farcinica In Complex With Testosterone
Organism: Nocardia farcinica
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2014-03-05 Classification: oxidoreductase/substrate Ligands: HEM, MG, TES, FMT, K |
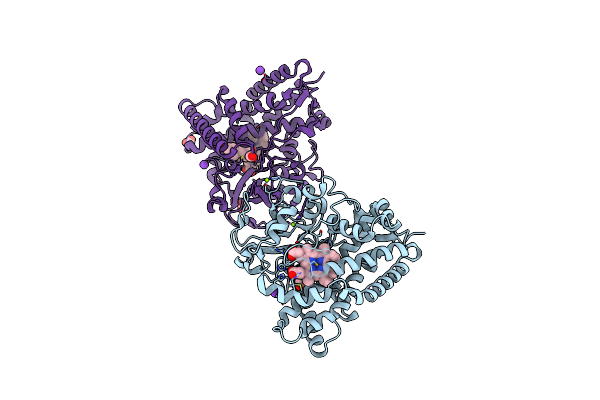 |
The 2.2 A Crystal Structure Of Cyp154C5 From Nocardia Farcinica In Complex With Androstenedione
Organism: Nocardia farcinica
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2014-03-05 Classification: oxidoreductase/substrate Ligands: ASD, HEM, MG, FMT, K |

