Search Count: 15
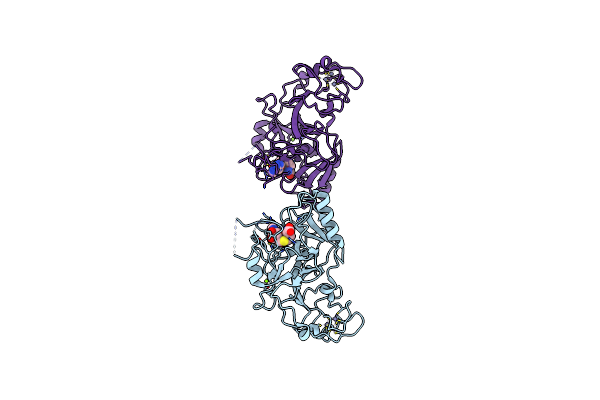 |
Organism: Schizosaccharomyces pombe (strain 972 / atcc 24843)
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2021-05-26 Classification: TRANSFERASE Ligands: ZN, SAH, MG |
 |
Crystal Structure Of The Chp2 Chromoshadow Domain In Complex With N-Terminal Domain Of Chromatin Remodeler Mit1
Organism: Schizosaccharomyces pombe
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2019-03-27 Classification: REPLICATION Ligands: HEZ |
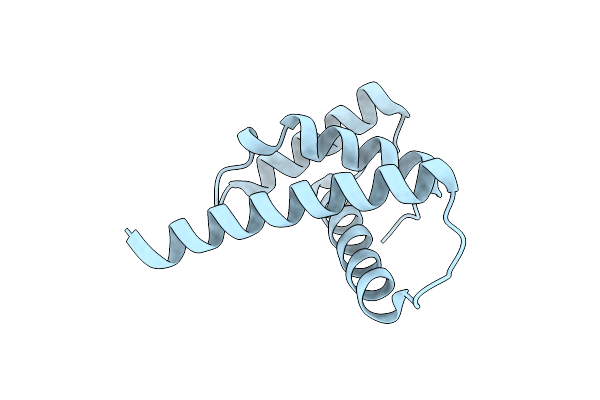 |
Organism: Schizosaccharomyces pombe (strain 972 / atcc 24843)
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2018-11-28 Classification: DNA BINDING PROTEIN |
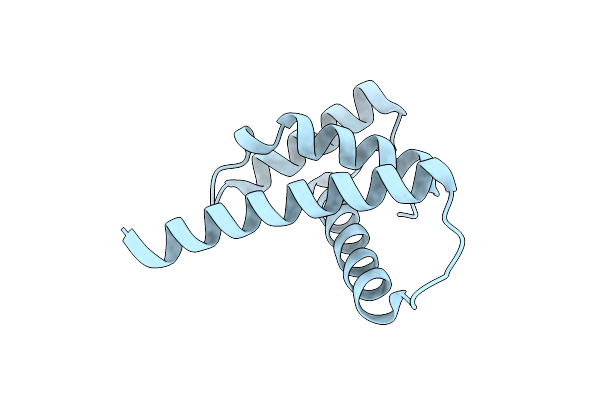 |
Organism: Schizosaccharomyces pombe (strain 972 / atcc 24843)
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2018-11-28 Classification: DNA BINDING PROTEIN |
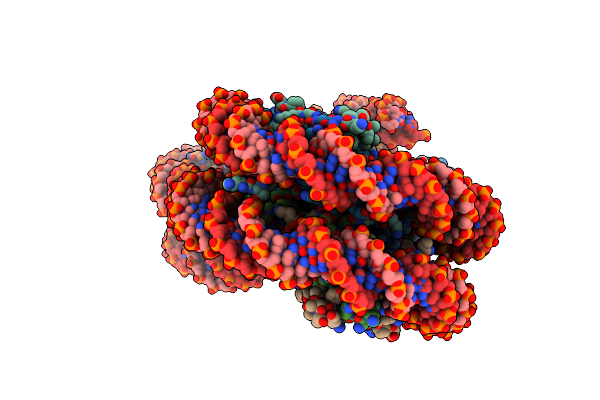 |
Organism: Synthetic construct, Xenopus laevis
Method: X-RAY DIFFRACTION Resolution:6.72 Å Release Date: 2017-10-11 Classification: GENE REGULATION |
 |
Organism: Xenopus laevis, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:5.77 Å Release Date: 2017-09-20 Classification: GENE REGULATION Ligands: CL |
 |
Crystal Structure Of The C-Terminal Domain Of The Mit1 Nucleosome Remodeler In Complex With Clr1
Organism: Schizosaccharomyces pombe (strain 972 / atcc 24843), Schizosaccharomyces pombe
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2016-04-20 Classification: TRANSCRIPTION Ligands: ZN, CL |
 |
Organism: Schizosaccharomyces pombe 972h-, Schizosaccharomyces pombe (strain 972 / atcc 24843)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2016-04-20 Classification: TRANSCRIPTION Ligands: ZN, CL |
 |
Organism: Schizosaccharomyces pombe 972h-
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2016-04-20 Classification: TRANSCRIPTION Ligands: ZN, K, NO3, EDO, SO4, MG |
 |
Organism: Schizosaccharomyces pombe
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2014-01-22 Classification: GENE REGULATION Ligands: EDO, CL |
 |
Organism: Saccharomyces cerevisiae, Schizosaccharomyces pombe, Schizosaccharomyces pombe (strain 972 / atcc 24843)
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2011-11-16 Classification: GENE REGULATION/PROTEIN BINDING Ligands: CL, K |
 |
Organism: Schizosaccharomyces pombe
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2009-04-21 Classification: NUCLEAR PROTEIN Ligands: ACY, ZN |
 |
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2006-02-09 Classification: CELL DIVISION PROTEIN Ligands: ZN, ADP, MG |
 |
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2006-02-09 Classification: HYDROLASE Ligands: ZN, ADP, MG |
 |
Organism: Xenopus laevis
Method: X-RAY DIFFRACTION Resolution:9.00 Å Release Date: 2005-07-12 Classification: STRUCTURAL PROTEIN/DNA |

