Search Count: 38
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-11-29 Classification: PROTEIN FIBRIL |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-11-29 Classification: PROTEIN FIBRIL |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-11-29 Classification: PROTEIN FIBRIL |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-11-29 Classification: PROTEIN FIBRIL |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-11-29 Classification: PROTEIN FIBRIL |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-11-29 Classification: PROTEIN FIBRIL |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-11-29 Classification: PROTEIN FIBRIL |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-11-29 Classification: PROTEIN FIBRIL |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-11-29 Classification: PROTEIN FIBRIL |
 |
Organism: Clostridium pasteurianum
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2023-10-25 Classification: OXIDOREDUCTASE Ligands: 402, SF4, FES, MG, GOL, CL |
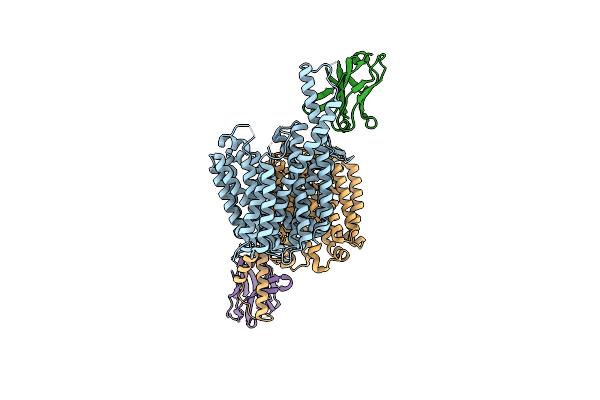 |
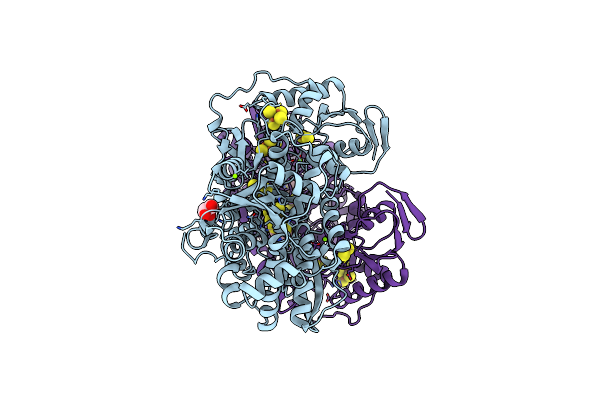
Outward-Open Conformation Of A Major Facilitator Superfamily (Mfs) Transporter Mhas2168, A Homologue Of Rv1410 From M. Tuberculosis, In Complex With An Alpaca Nanobody
Organism: Mycolicibacterium hassiacum dsm 44199, Vicugna pacos
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2023-10-18 Classification: TRANSPORT PROTEIN |
 |
Organism: Clostridium pasteurianum
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2023-09-27 Classification: OXIDOREDUCTASE Ligands: 402, SF4, FES, MG, GOL |
 |
Sh3 Domain Solved By The Exact Solid-State Method From The Bruker Dynamics Center Using The Combined Correction Method With Pdb 2Nuz
Organism: Gallus gallus
Method: SOLID-STATE NMR Release Date: 2023-03-08 Classification: PEPTIDE BINDING PROTEIN |
 |
Sh3 Domain Solved By The Exact Solid-State Method From The Bruker Dynamics Center Using The Correction For Dipolar Truncation With Pdb 2Nuz
Organism: Gallus gallus
Method: SOLID-STATE NMR Release Date: 2023-03-08 Classification: PEPTIDE BINDING PROTEIN |
 |
Sh3 Domain Solved By The Exact Solid-State Method From The Bruker Dynamics Center Using The Correction Method For Spin-Diffusion With Pdb 2Nuz As Reference
Organism: Gallus gallus
Method: SOLID-STATE NMR Release Date: 2023-03-08 Classification: PEPTIDE BINDING PROTEIN |
 |
The Solution Structure Of The Triple Mutant Methyl-Cpg-Binding Domain From Mecp2
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2023-02-22 Classification: GENE REGULATION |
 |
The Solution Structure Of The Triple Mutant Methyl-Cpg-Binding Domain From Mecp2 That Binds To Asymmetrically Modified Dna
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2023-02-22 Classification: DNA BINDING PROTEIN |
 |
Nanodisc Reconstituted Msba In Complex With Nanobodies, Spin-Labeled At Position A60C
Organism: Escherichia coli k-12, Vicugna pacos
Method: ELECTRON MICROSCOPY Release Date: 2022-08-24 Classification: MEMBRANE PROTEIN Ligands: EIW, 88T, GD |
 |
Amp-Pnp Bound Nanodisc Reconstituted Msba With Nanobodies, Spin-Labeled At Position A60C
Organism: Escherichia coli k-12, Vicugna pacos
Method: ELECTRON MICROSCOPY Resolution:2.80 Å Release Date: 2022-08-24 Classification: MEMBRANE PROTEIN Ligands: ANP, MG, LMT, 3PE, 88T, GD |
 |
Amp-Pnp Bound Nanodisc Reconstituted Msba With Nanobodies, Spin-Labeled At Position T68C
Organism: Escherichia coli (strain k12), Vicugna pacos
Method: ELECTRON MICROSCOPY Release Date: 2022-08-24 Classification: MEMBRANE PROTEIN Ligands: ANP, MG, LMT, 88T, GD |

