Search Count: 71
 |
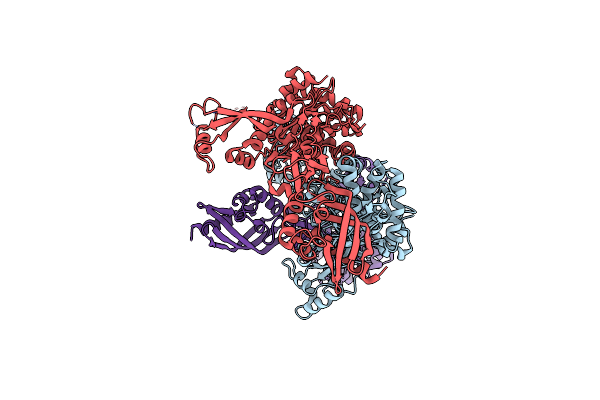
Crystal Structure Of Enterococcus Faecium D63R Penicillin-Binding Protein 5 (Pbp5Fm)
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2019-04-24 Classification: ANTIBIOTIC Ligands: RB6, SO4 |
 |
Crystal Structure Of Enterococcus Faecium D63R Penicillin-Binding Protein 5 (Pbp5Fm)
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2019-04-10 Classification: ANTIBIOTIC Ligands: SO4 |
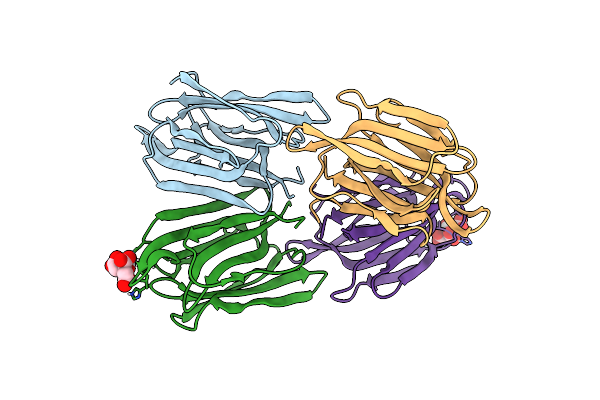 |
Structure Of Acmjrl, A Mannose Binding Jacalin Related Lectin From Ananas Comosus.
Organism: Ananas comosus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-08-15 Classification: SUGAR BINDING PROTEIN Ligands: CIT |
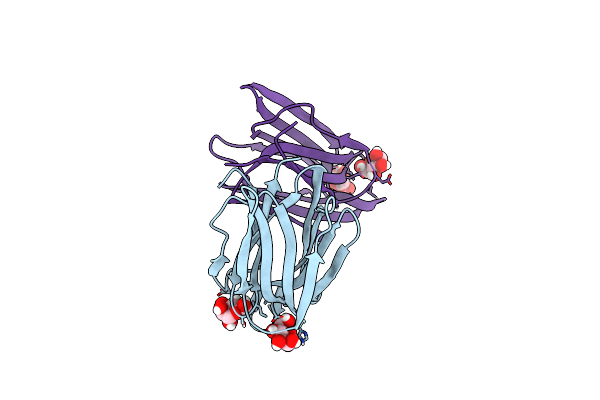 |
Structure Of Acmjrl, A Mannose Binding Jacalin Related Lectin From Ananas Comosus, In Complex With Mannose.
Organism: Ananas comosus
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2018-08-15 Classification: SUGAR BINDING PROTEIN Ligands: MAN |
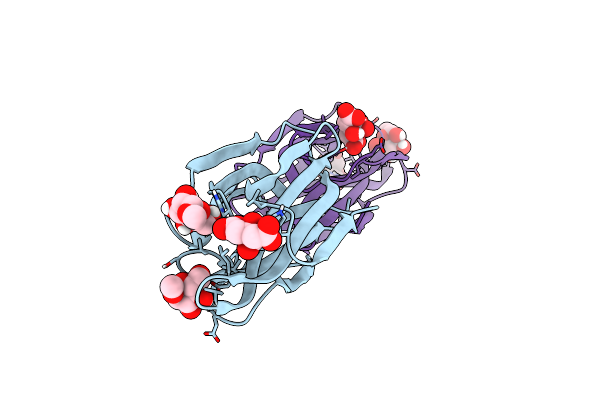 |
Structure Of Acmjrl, A Mannose Binding Jacalin Related Lectin From Ananas Comosus, In Complex With Methyl-Mannose.
Organism: Ananas comosus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-08-15 Classification: SUGAR BINDING PROTEIN Ligands: MMA, CIT |
 |
Structure Of A Single Domain Camelid Antibody Fragment Cab-G10S In Complex With The Blap Beta-Lactamase From Bacillus Licheniformis
Organism: Bacillus licheniformis, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.56 Å Release Date: 2017-11-15 Classification: HYDROLASE Ligands: ACT |
 |
Crystal Structure Of The First Transmembrane Pap2 Type Phosphatidylglycerolphosphate Phosphatase From Bacillus Subtilis
Organism: Bacillus subtilis (strain 168)
Method: X-RAY DIFFRACTION Release Date: 2017-02-22 Classification: HYDROLASE Ligands: WO4, OLC, UNL |
 |
Crystal Structure Of The Class B3 Di-Zinc Metallo-Beta-Lactamase Lra- 12 From An Alaskan Soil Metagenome.
Organism: Uncultured bacterium blr12
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2015-09-16 Classification: HYDROLASE Ligands: ZN, SO4, CO |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2014-08-13 Classification: ISOMERASE Ligands: GOL, CIT |
 |
Crystal Structure Of Galactose Mutarotase Galm From Bacillus Subtilis With Trehalose
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2014-08-13 Classification: ISOMERASE Ligands: PGE, CIT, ACT |
 |
Crystal Structure Of Galactose Mutarotase Galm From Bacillus Subtilis In Complex With Maltose
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2014-08-13 Classification: ISOMERASE Ligands: CIT |
 |
Crystal Structure Of Galactose Mutarotase Galm From Bacillus Subtilis In Complex With Maltose And Trehalose
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2014-08-13 Classification: ISOMERASE Ligands: GOL, CIT |
 |
Organism: Citrobacter freundii
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2014-05-21 Classification: HYDROLASE |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2014-05-07 Classification: TRANSFERASE Ligands: SO4, CXS, GOL, EDO, CL |
 |
Crystal Structure Of E. Coli Penicillin Binding Protein 3, Domain V88- S165
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2014-05-07 Classification: TRANSFERASE Ligands: SO4, TRS |
 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2014-01-08 Classification: HYDROLASE Ligands: TBE, SO4 |
 |
Crystal Structure Of The Class A Extended-Spectrum Beta-Lactamase Ctx- M-96, A Natural D240G Mutant Derived From Ctx-M-12
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2014-01-08 Classification: HYDROLASE |
 |
Crystal Structure Of A Complex Between Actinomadura R39 Dd-Peptidase And A Sulfonamide Boronate Inhibitor
Organism: Actinomadura sp r39
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2013-08-21 Classification: HYDROLASE Ligands: HQZ, SO4, MG |
 |
Crystal Structure Of A Complex Between Actinomadura R39 Dd-Peptidase And A Sulfonamide Boronate Inhibitor
Organism: Actinomadura sp. r39
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2013-08-21 Classification: HYDROLASE Ligands: BSF, MG, SO4 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2013-08-21 Classification: HYDROLASE Ligands: ZN, NA |

