Search Count: 18
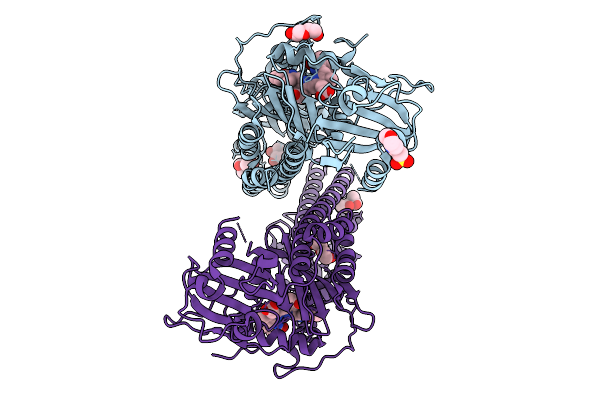 |
Crystal Structure Of The Photosensory Core Module (Pcm) Of A Cyano-Phenylalanine Mutant Ocnf205 Of The Bathy Phytochrome Agp2 From Agrobacterium Fabrum In The Pfr State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2025-10-08 Classification: SIGNALING PROTEIN Ligands: EL5, MPD, PEG, MES, CL |
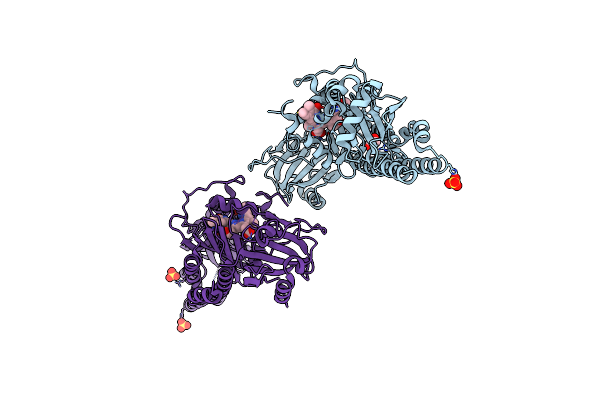 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In Its Pfr State (I0A).
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, CL, EDO |
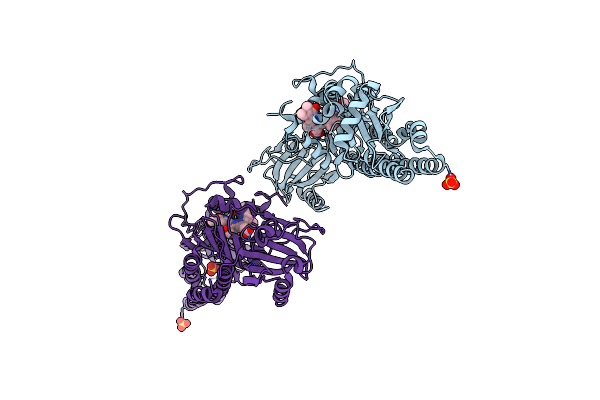 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In Its Pfr State (I0B).
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4 |
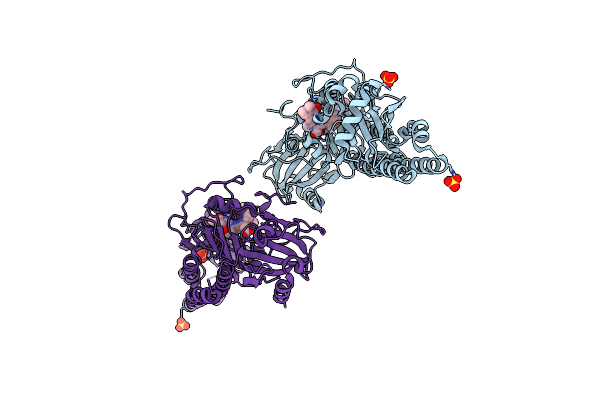 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I1 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Resolution:2.54 Å Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4 |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I2 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, PGE, PEG, CL |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I3 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, GOL, PEG |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I4 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, CL, PEG |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I5 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, CL |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I6 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, CL |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I7 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, PEG |
 |
Crystal Structure Of The Photosensory Core Module (Pcm) Of A Cyano-Phenylalanine Mutant Ocnf165 Of The Bathy Phytochrome Agp2 From Agrobacterium Fabrum In The Pfr State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-11-27 Classification: SIGNALING PROTEIN Ligands: P6G, MPD, PEG, EL5, CL |
 |
Crystal Structure Of The Photosensory Core Module (Pcm) Of A Cyano-Phenylalanine Mutant Ocnf192 Of The Bathy Phytochrome Agp2 From Agrobacterium Fabrum In The Pfr State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2024-11-27 Classification: SIGNALING PROTEIN Ligands: PEG, MPD, EL5, CL |
 |
The Surface-Engineered Photosensory Module (Pas-Gaf-Phy) Of The Bacterial Phytochrome Agp1 (Atbphp1) In The Pr Form With Parallel Dimer Formation
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2024-03-06 Classification: SIGNALING PROTEIN Ligands: V8U, MG |
 |
The Photosensory Core Module (Pas-Gaf-Phy) Of The Bacterial Phytochrome Agp1 (Atbphp1) Locked In A Pr-Like State
Organism: Agrobacterium fabrum (strain c58 / atcc 33970)
Method: X-RAY DIFFRACTION Resolution:3.11 Å Release Date: 2020-04-01 Classification: SIGNALING PROTEIN Ligands: JQ2, CA |
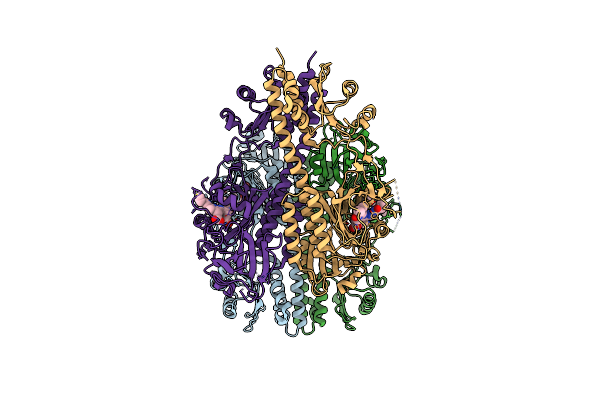 |
Crystallographic Superstructure Of The Photosensory Core Module (Pas-Gaf-Phy) Of The Bacterial Phytochrome Agp1 (Atbphp1) Locked In A Pr-Like State
Organism: Agrobacterium fabrum (strain c58 / atcc 33970)
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2020-04-01 Classification: SIGNALING PROTEIN Ligands: JQ2 |
 |
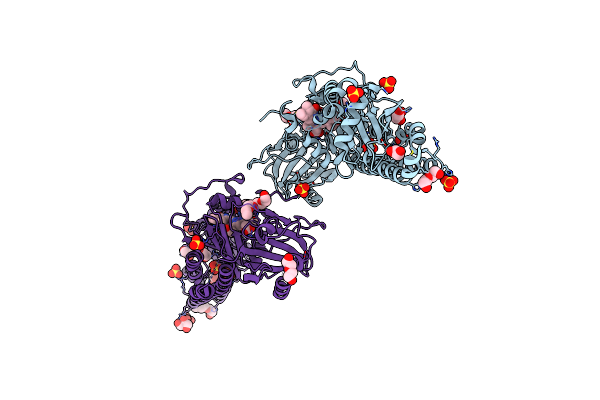
Crystal Structure Of The Photosensory Core Module (Pcm) Of A Bathy Phytochrome From Agrobacterium Fabrum In The Pfr State.
Organism: Agrobacterium fabrum (strain c58 / atcc 33970)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2018-11-28 Classification: SIGNALING PROTEIN Ligands: EL5 |
 |
Crystal Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In Its Pfr State.
Organism: Agrobacterium fabrum (strain c58 / atcc 33970)
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2018-11-28 Classification: FLUORESCENT PROTEIN Ligands: EL5, SO4, GOL, PEG, ETE, PG0 |
 |
Crystal Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In Its Functional Meta-F Intermediate State.
Organism: Agrobacterium fabrum (strain c58 / atcc 33970)
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2018-11-28 Classification: FLUORESCENT PROTEIN Ligands: EL5, SO4, CL, GOL, PEG, ETE, PG0 |

