Search Count: 16
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2019-06-05 Classification: SUGAR BINDING PROTEIN Ligands: GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2019-06-05 Classification: SUGAR BINDING PROTEIN Ligands: GOL |
 |
X-Ray Structure Of A Non-Autocrystallizing Galectin-10 Variant, Gal10-Tyr69Glu
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-06-05 Classification: SUGAR BINDING PROTEIN Ligands: PGE, PG4, P6G |
 |
Structure Of Galectin-10 In Complex With The Fab Fragment Of A Charcot-Leyden Crystal Solubilizing Antibody, 6F5
Organism: Homo sapiens, Lama glama
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2019-06-05 Classification: SUGAR BINDING PROTEIN Ligands: CA, PGE |
 |
Structure Of Galectin-10 In Complex With The Fab Fragment Of A Charcot-Leyden Crystal Solubilizing Antibody, 1D11
Organism: Homo sapiens, Lama glama
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-06-05 Classification: SUGAR BINDING PROTEIN Ligands: PG4, ACT |
 |
Structure Of Galectin-10 In Complex With The Fab Fragment Of A Charcot-Leyden Crystal Solubilizing Antibody, 4E8
Organism: Homo sapiens, Lama glama
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2019-06-05 Classification: SUGAR BINDING PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2019-06-05 Classification: IMMUNE SYSTEM Ligands: RIP |
 |
Organism: Homo sapiens, Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2018-11-07 Classification: IMMUNE SYSTEM |
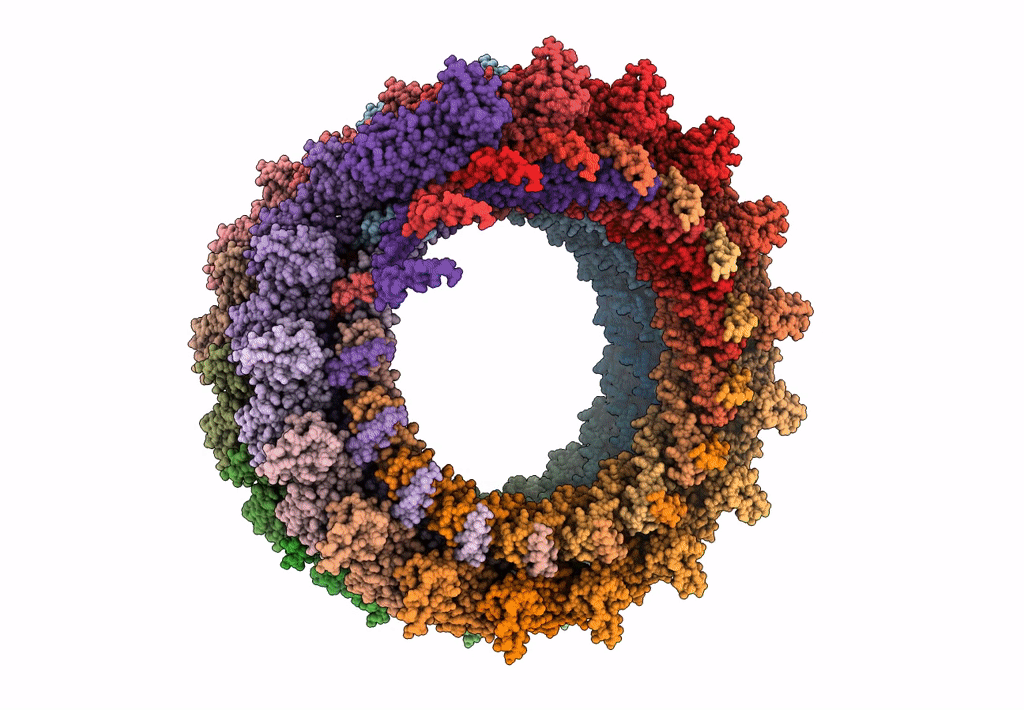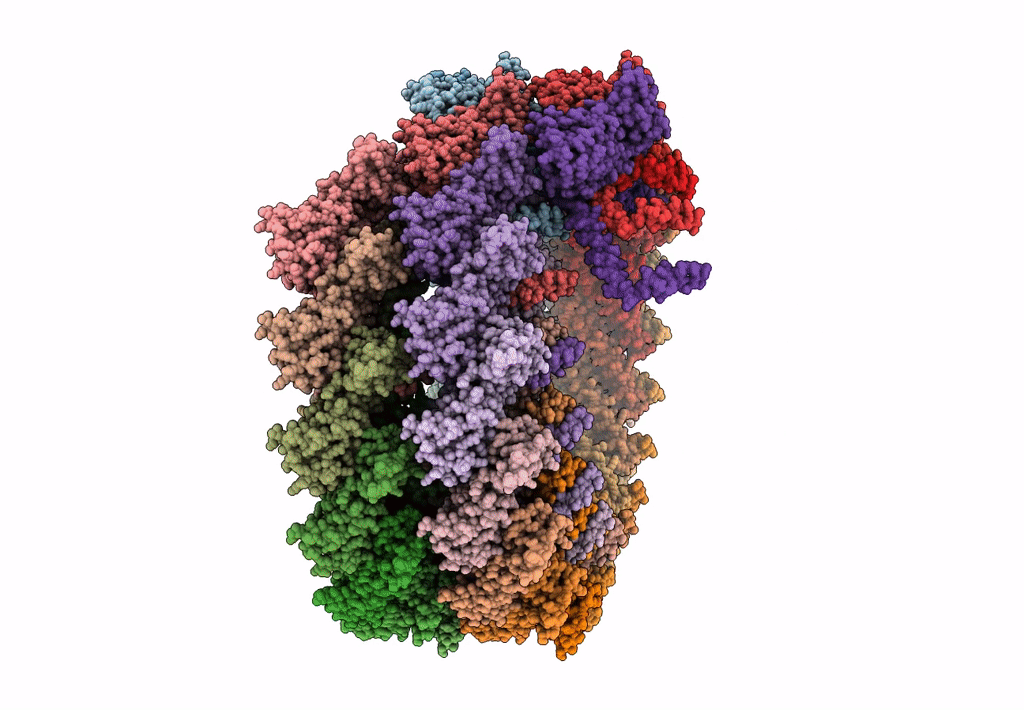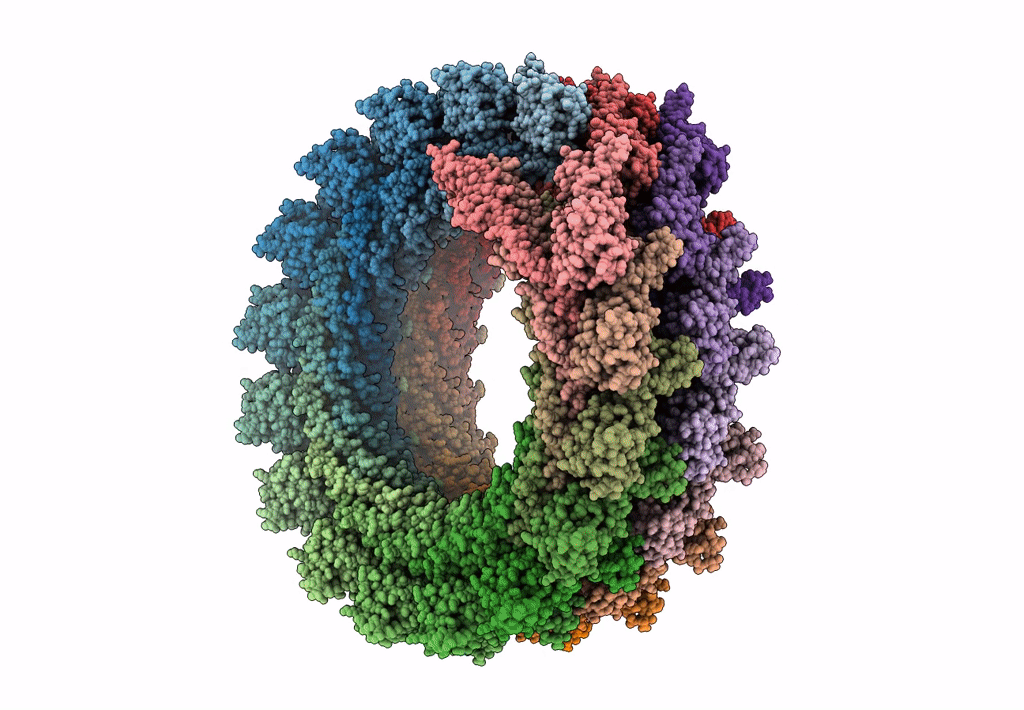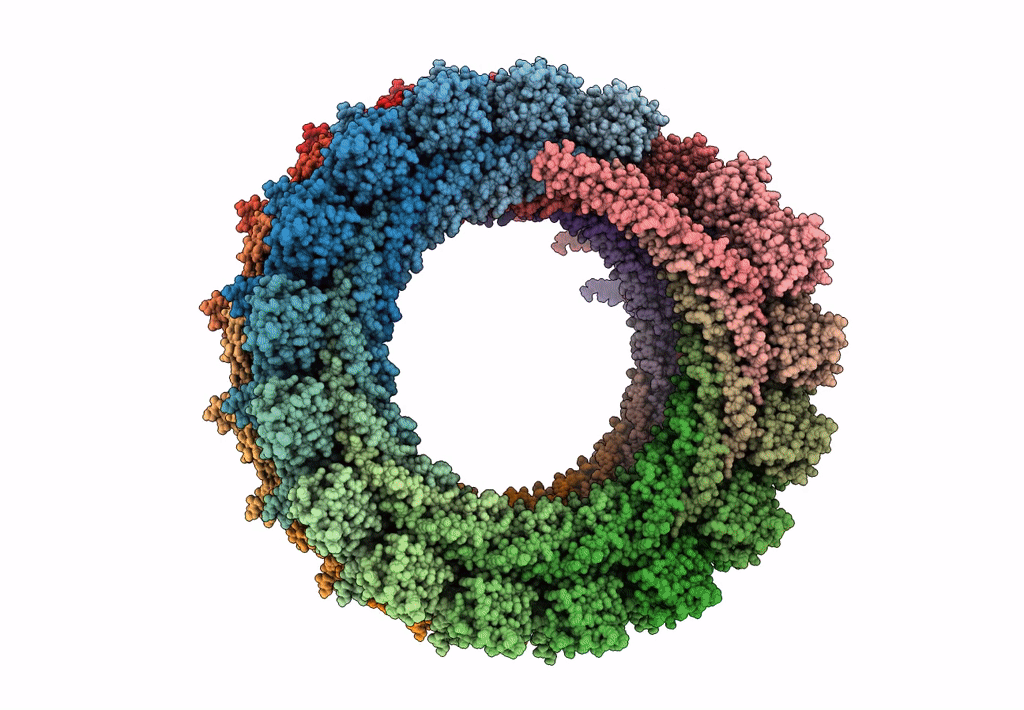 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2015-12-16 Classification: LIPID BINDING PROTEIN |
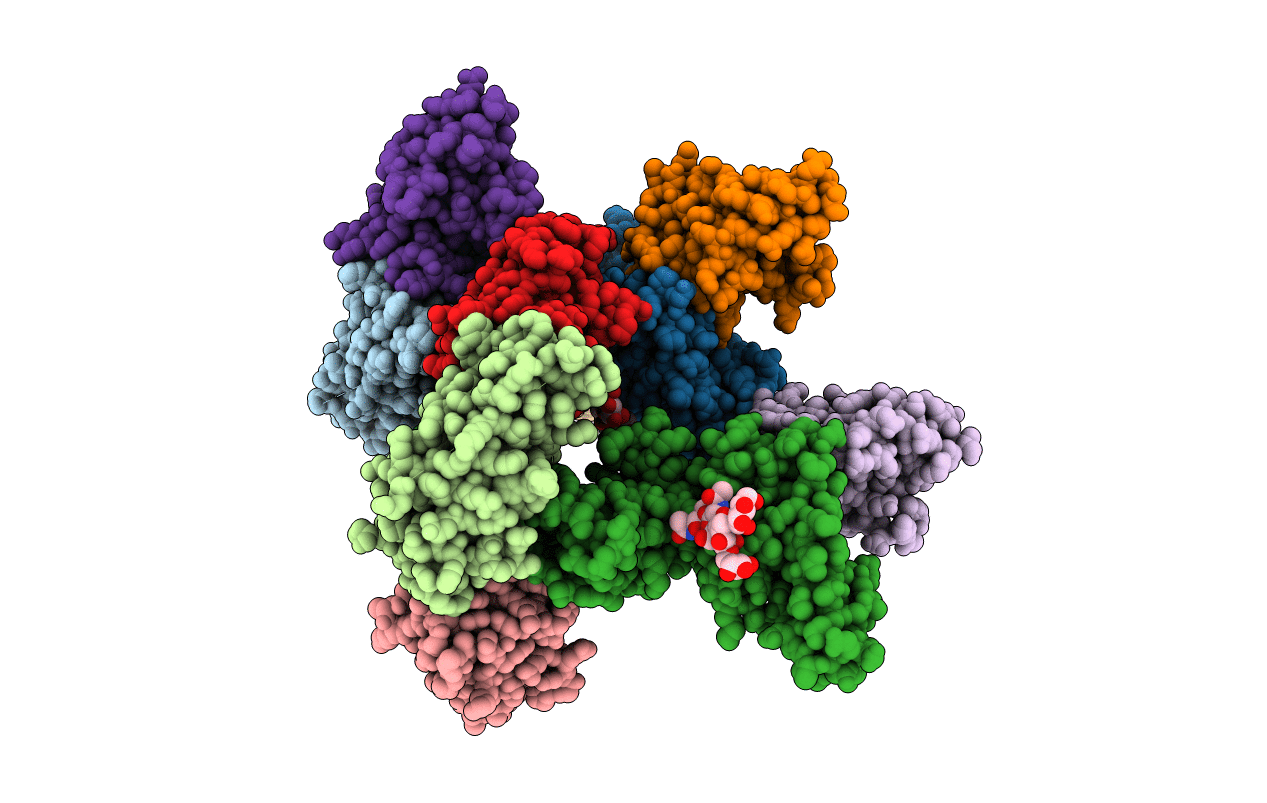 |
Organism: Homo sapiens, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2014-02-19 Classification: IMMUNE SYSTEM |
 |
The Twinned 3.35A Structure Of S. Aureus Gyrase Complex With Ciprofloxacin And Dna
Organism: Staphylococcus aureus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.35 Å Release Date: 2010-08-25 Classification: ISOMERASE/DNA/ANTIBIOTIC Ligands: MN, CPF |
 |
The 3.1A Crystal Structure Of The Catalytic Core (B'A' Region) Of Staphylococcus Aureus Dna Gyrase
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2010-08-04 Classification: ISOMERASE Ligands: CA |
 |
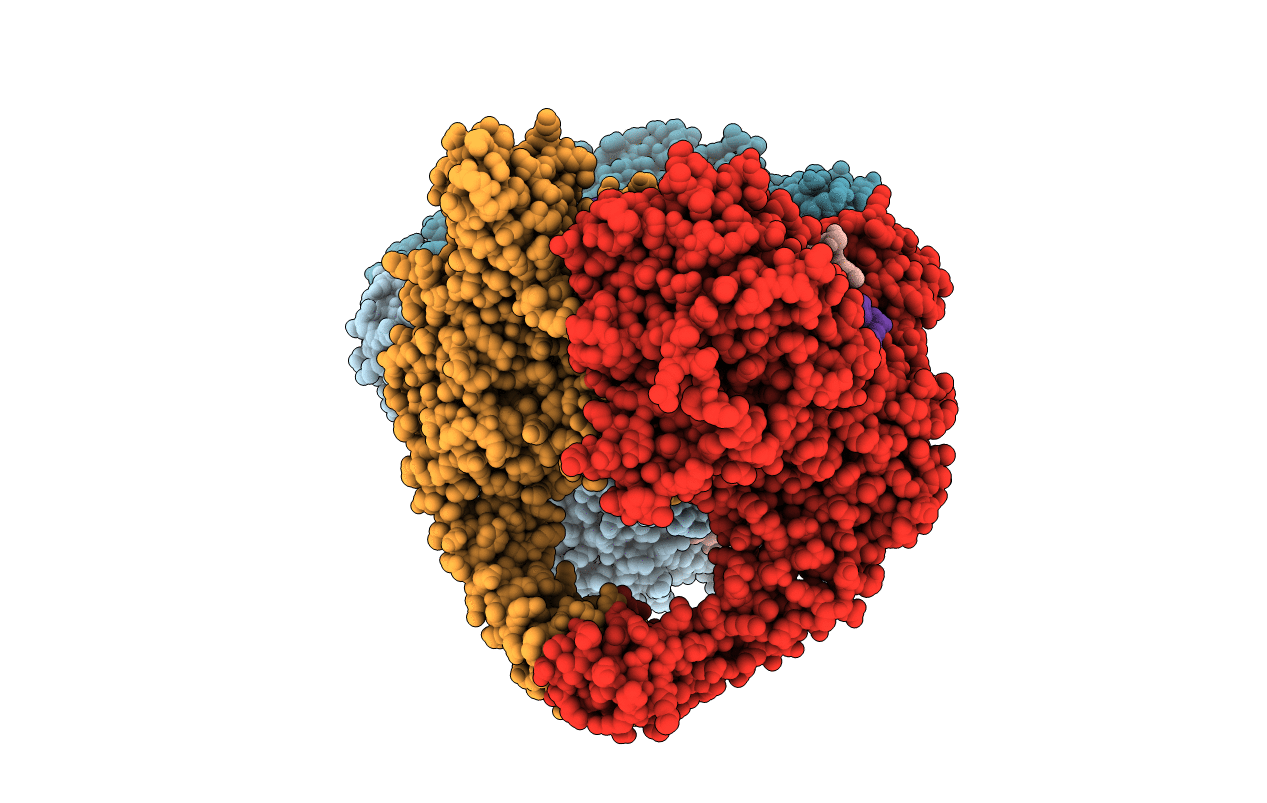
The 2.98A Crystal Structure Of The Catalytic Core (B'A' Region) Of Staphylococcus Aureus Dna Gyrase
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.98 Å Release Date: 2010-08-04 Classification: ISOMERASE Ligands: HOH |
 |
The 3.5A Crystal Structure Of The Catalytic Core (B'A' Region) Of Staphylococcus Aureus Dna Gyrase Complexed With Gsk299423 And Dna
Organism: Staphylococcus aureus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2010-08-04 Classification: ISOMERASE Ligands: RXV |
 |
The 2.1A Crystal Structure Of S. Aureus Gyrase Complex With Gsk299423 And Dna
Organism: Staphylococcus aureus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2010-08-04 Classification: ISOMERASE Ligands: MN, RXV |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2007-04-24 Classification: TRANSCRIPTION Ligands: 735 |

