Search Count: 24
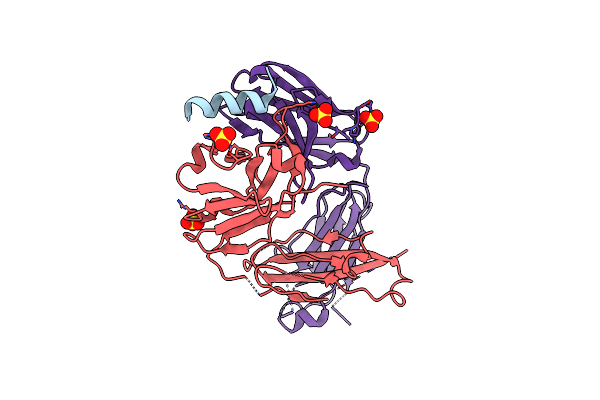 |
S2P6 Fab Fragment Bound To The Sars-Cov/Sars-Cov-2 Spike Stem Helix Peptide
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.67 Å Release Date: 2021-08-11 Classification: ANTIVIRAL PROTEIN, IMMUNE SYSTEM Ligands: SO4 |
 |
Organism: Mus musculus, Human coronavirus oc43
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-05-26 Classification: ANTIVIRAL PROTEIN, IMMUNE SYSTEM Ligands: GOL |
 |
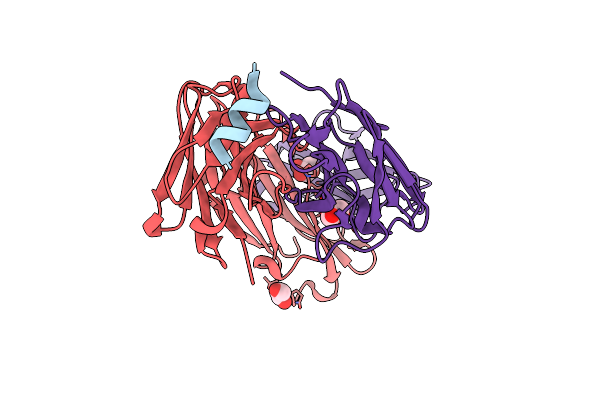
Organism: Mus musculus, Bat coronavirus hku4
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2021-05-26 Classification: ANTIVIRAL PROTEIN, IMMUNE SYSTEM Ligands: GOL |
 |
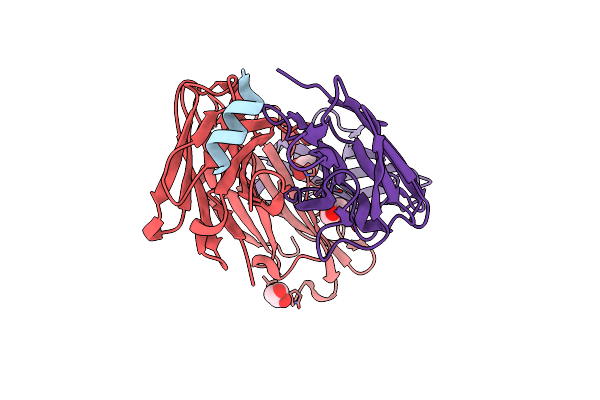
Organism: Mus musculus, Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2021-05-26 Classification: ANTIVIRAL PROTEIN, IMMUNE SYSTEM Ligands: GOL |
 |
Organism: Mus musculus, Middle east respiratory syndrome-related coronavirus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2021-05-26 Classification: ANTIVIRAL PROTEIN, IMMUNE SYSTEM Ligands: GOL |
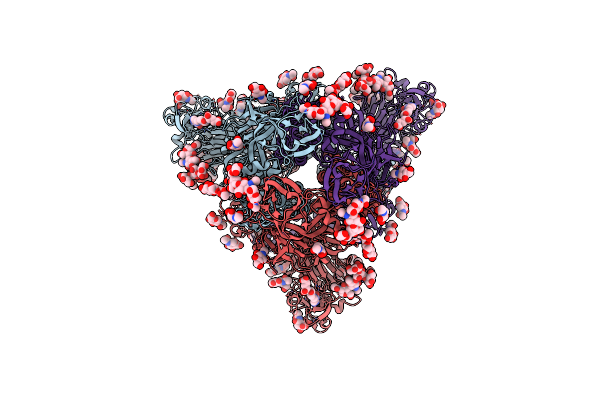 |
Mers-Cov S Bound To The Broadly Neutralizing B6 Fab Fragment (C3 Refinement)
Organism: Middle east respiratory syndrome-related coronavirus
Method: ELECTRON MICROSCOPY Resolution:2.50 Å Release Date: 2021-05-26 Classification: VIRAL PROTEIN Ligands: FOL, NAG, SIA |
 |
Organism: Human betacoronavirus 2c emc/2012
Method: ELECTRON MICROSCOPY Resolution:2.50 Å Release Date: 2019-12-11 Classification: VIRAL PROTEIN Ligands: NAG, FOL, SIA |
 |
Organism: Human betacoronavirus 2c emc/2012
Method: ELECTRON MICROSCOPY Release Date: 2019-12-11 Classification: VIRAL PROTEIN Ligands: NAG, FOL |
 |
Organism: Human betacoronavirus 2c emc/2012
Method: ELECTRON MICROSCOPY Release Date: 2019-12-11 Classification: VIRAL PROTEIN Ligands: NAG, FOL |
 |
Organism: Human betacoronavirus 2c emc/2012
Method: ELECTRON MICROSCOPY Resolution:2.90 Å Release Date: 2019-12-11 Classification: VIRAL PROTEIN Ligands: NAG, FOL |
 |
Crystal Structure Of A Fimh*Dsg Complex From E.Coli F18 With Bound Trimannose
Organism: Escherichia coli f18+, Escherichia coli 536
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-01-16 Classification: CELL ADHESION Ligands: PEG, 1PE |
 |
Crystal Structure Of The Fimh Lectin Domain From E.Coli F18 In Complex With Trimannose
Organism: Escherichia coli f18+
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2019-01-16 Classification: CELL ADHESION Ligands: CA |
 |
Crystal Structure Of The Fimh Lectin Domain From E.Coli K12 In Complex With The Dimannoside Man(Alpha1-2)Man
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2019-01-16 Classification: CELL ADHESION Ligands: SO4 |
 |
Crystal Structure Of The Fimh Lectin Domain From E.Coli K12 In Complex With The Dimannoside Man(Alpha1-6)Man
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-01-16 Classification: CELL ADHESION |
 |
Crystal Structure Of A Fimh*Dsg Complex From E.Coli F18 With Bound Dimannoside Man(Alpha1-3)Man In Space Group P21
Organism: Escherichia coli f18+, Escherichia coli 536
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2019-01-16 Classification: CELL ADHESION |
 |
Crystal Structure Of A Fimh*Dsg Complex From E.Coli F18 With Bound Dimannoside Man(Alpha1-3)Man In Space Group P213
Organism: Escherichia coli f18+, Escherichia coli 536
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2019-01-16 Classification: CELL ADHESION Ligands: SO4 |
 |
Organism: Escherichia coli, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2017-11-08 Classification: TRANSPORT PROTEIN Ligands: GOL |
 |
Crystal Structure Of The Fimh Lectin Domain From E.Coli K12 In Complex With Heptyl Alpha-D-Mannopyrannoside
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2016-01-27 Classification: CELL ADHESION Ligands: KGM |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.14 Å Release Date: 2016-01-27 Classification: CELL ADHESION |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.54 Å Release Date: 2016-01-27 Classification: CELL ADHESION |

