Search Count: 39
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-06-05 Classification: CELL ADHESION Ligands: MG, CA |
 |
Crystal Structure Of L-Methionine Decarboxylase From Streptomyces Sp.590 (Internal Aldimine Form).
Organism: Streptomyces sp. 590 ki-2014
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-01-27 Classification: LYASE |
 |
Crystal Structure Of L-Methionine Decarboxylase Q64A Mutant From Streptomyces Sp.590 In Complexed With L- Methionine Methyl Ester (Geminal Diamine Form).
Organism: Streptomyces sp. 590 ki-2014
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2021-01-27 Classification: LYASE Ligands: G03 |
 |
Crystal Structure Of L-Methionine Decarboxylase From Streptomyces Sp.590 In Complexed With L- Methionine Methyl Ester (External Aldimine Form).
Organism: Streptomyces sp. 590 ki-2014
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2021-01-27 Classification: LYASE Ligands: G06 |
 |
Crystal Structure Of L-Methionine Decarboxylase From Streptomyces Sp.590 In Complexed With 3-Methlythiopropylamine (External Aldimine Form).
Organism: Streptomyces sp. 590 ki-2014
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2021-01-27 Classification: LYASE Ligands: G0C |
 |
Crystal Structure Of L-Methionine Decarboxylase From Streptomyces Sp.590 In Complexed With 3-Methlythiopropylamine (Geminal Diamine Form).
Organism: Streptomyces sp. 590 ki-2014
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-01-27 Classification: LYASE Ligands: G0F |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-09-11 Classification: MEMBRANE PROTEIN Ligands: SO4, ACT, FMN, ORO, 9AU |
 |
Organism: Eimeria tenella
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2019-08-28 Classification: MEMBRANE PROTEIN Ligands: FMN, ORO |
 |
Organism: Eimeria tenella
Method: X-RAY DIFFRACTION Resolution:3.65 Å Release Date: 2019-08-28 Classification: MEMBRANE PROTEIN Ligands: FMN, ORO, 9AU |
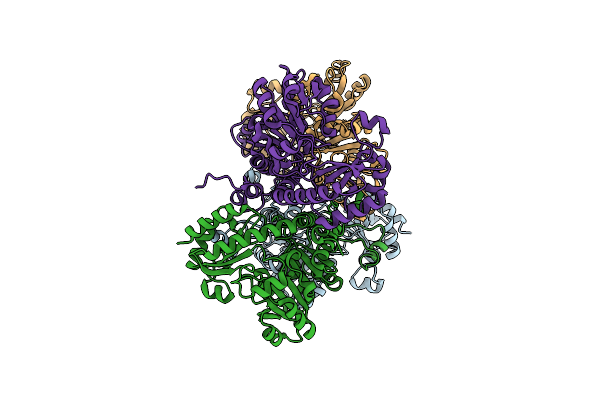 |
Crystal Structure Of Pseudomonas Putida Methionine Gamma-Lyase Wild Type Without Sulfate Ion
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2017-04-12 Classification: LYASE |
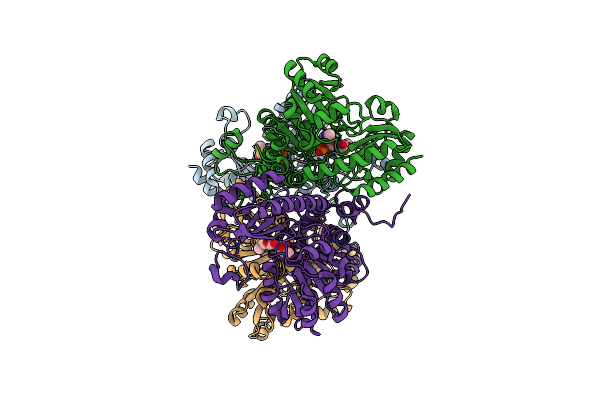 |
Crystal Structure Of Pseudomonas Putida Methionine Gamma-Lyase Wild Type With L-Methionine Intermediates
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2017-04-12 Classification: LYASE Ligands: 3LM |
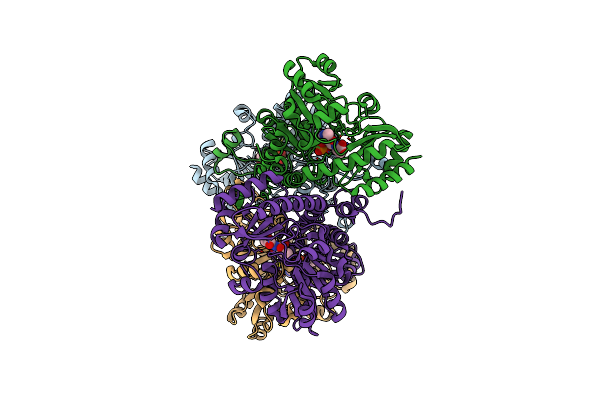 |
Crystal Structure Of Pseudomonas Putida Methionine Gamma-Lyase Wild Type With L-Homocysteine Intermediates
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-04-12 Classification: LYASE Ligands: 4LM, H2S |
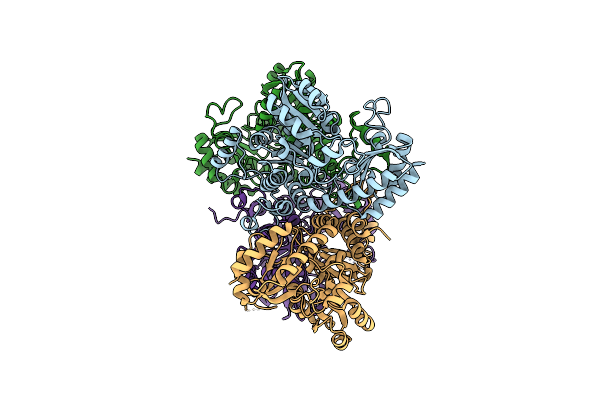 |
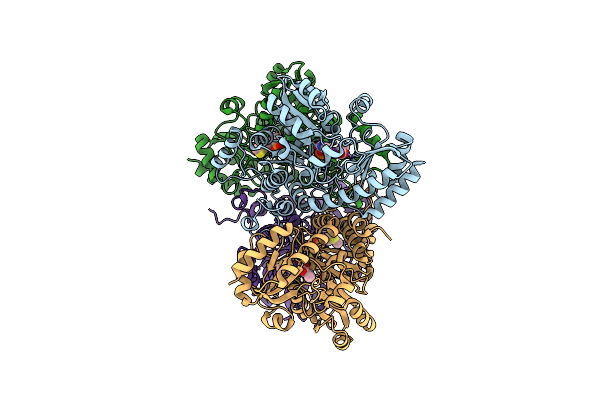
Crystal Structure Of Pseudomonas Putida Methionine Gamma-Lyase C116H Mutant Without Sulfate Ion
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2017-04-12 Classification: LYASE |
 |
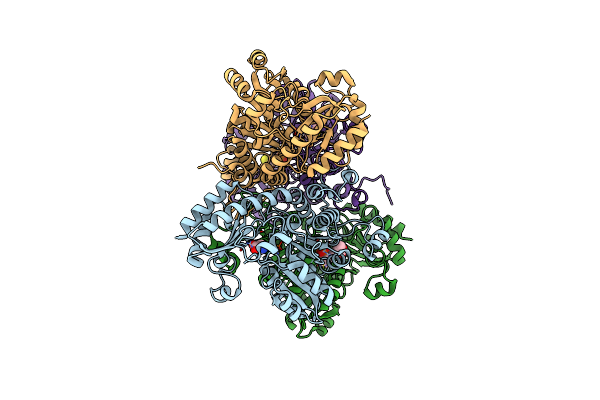
Crystal Structure Of Pseudomonas Putida Methionine Gamma-Lyase C116H Mutant With L-Methionine Intermediates
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-04-12 Classification: LYASE Ligands: 3LM |
 |
Crystal Structure Of Pseudomonas Putida Methionine Gamma-Lyase C116H Mutant With L-Homocysteine Intermediates.
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2017-04-12 Classification: LYASE Ligands: HCS, 4LM, H2S, 7XF |
 |
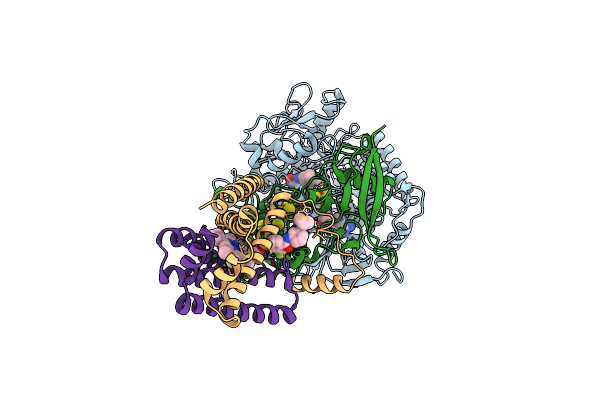
Crystal Structure Of Mitochondrial Rhodoquinol-Fumarate Reductase From Ascaris Suum With Ubiquinone-1
Organism: Ascaris suum
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2016-06-22 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: MLI, FAD, FES, SF4, F3S, HEM, UQ1, EPH |
 |
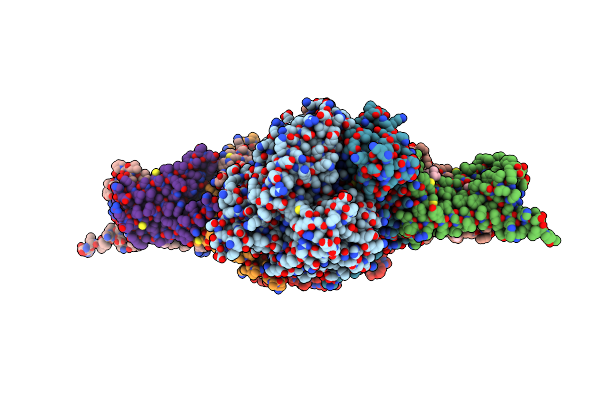
Crystal Structure Of Porcine Heart Mitochondrial Complex Ii Bound With Flutolanil
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2016-03-02 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: FAD, FES, SF4, F3S, HEM, FTN |
 |
Crystal Structure Of Mitochondrial Rhodoquinol-Fumarate Reductase From Ascaris Suum With The Specific Inhibitor Nn23
Organism: Ascaris suum
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2015-08-05 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: MLI, FAD, FES, SF4, F3S, E23, HEM, EPH |
 |
Crystal Structure Of Mitochondrial Rhodoquinol-Fumarate Reductase From Ascaris Suum With N-[(2,4-Dichlorophenyl)Methyl]-2-(Trifluoromethyl)Benzamide
Organism: Ascaris suum
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2015-08-05 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: MLI, FAD, FES, SF4, F3S, E24, HEM, EPH |
 |
Crystal Structure Of Mitochondrial Rhodoquinol-Fumarate Reductase From Ascaris Suum With 2-Iodo-N-[3-(1-Methylethoxy)Phenyl]Benzamide
Organism: Ascaris suum
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2015-08-05 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: MLI, FAD, FES, SF4, F3S, HEM, 12J, EPH |

