Search Count: 15
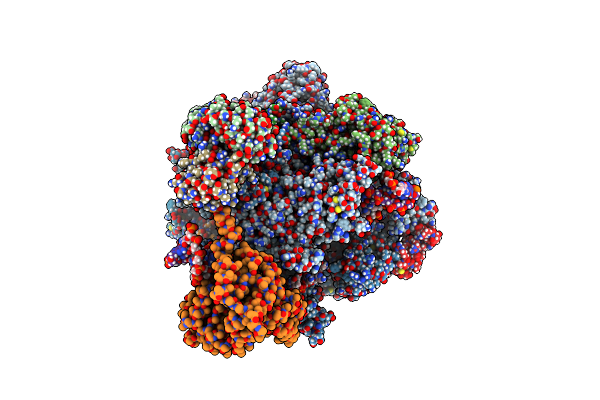 |
Cryo-Em Structure Of Cpd-Stalled Pol Ii In Complex With Rad26 (Engaged State)
Organism: Saccharomyces cerevisiae, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-01-24 Classification: TRANSCRIPTION/DNA/RNA Ligands: ZN, MG |
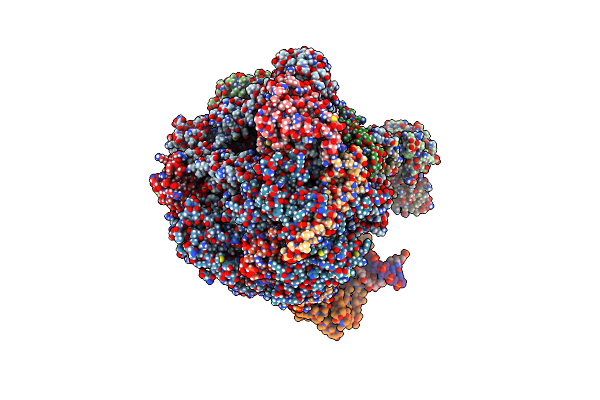 |
Organism: Saccharomyces cerevisiae, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-01-24 Classification: TRANSCRIPTION/DNA/RNA Ligands: ZN, MG |
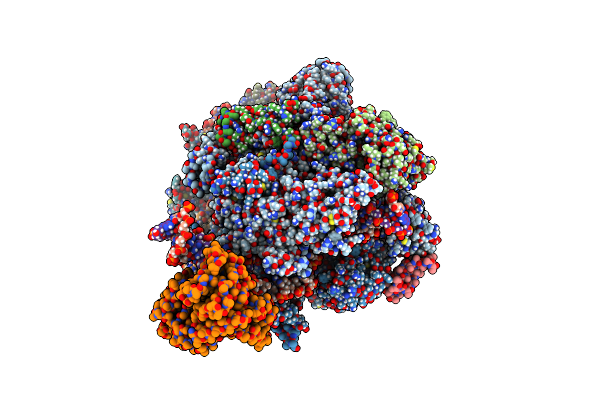 |
Organism: Saccharomyces cerevisiae, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-01-24 Classification: TRANSCRIPTION/DNA/RNA Ligands: ZN, MG |
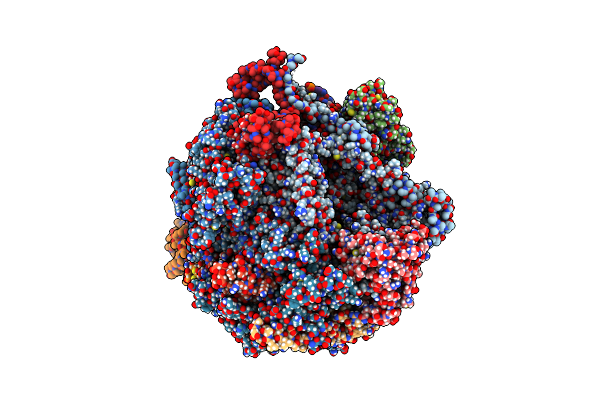 |
Organism: Saccharomyces cerevisiae, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-01-24 Classification: TRANSCRIPTION/DNA/RNA Ligands: ZN, MG |
 |
Organism: Synthetic construct, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-01-24 Classification: TRANSCRIPTION/DNA/RNA Ligands: ZN, MG |
 |
Organism: Synthetic construct, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-01-24 Classification: TRANSCRIPTION/DNA/RNA Ligands: ZN, MG |
 |
Organism: Synthetic construct, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-01-24 Classification: TRANSCRIPTION/DNA/RNA Ligands: ZN, MG |
 |
Cryo-Em Structure Of Cpd Lesion Containing Rna Polymerase Ii Elongation Complex With Rad26 And Elf1 (Closed State)
Organism: Saccharomyces cerevisiae, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-01-24 Classification: TRANSCRIPTION/DNA/RNA Ligands: ZN, MG |
 |
Organism: Mus musculus, Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2023-07-12 Classification: ENDOCYTOSIS Ligands: PIO |
 |
Structure Of M. Tuberculosis Cysq, A Pap Phosphatase - Ligand-Free Structure
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2015-11-11 Classification: HYDROLASE |
 |
Structure Of M. Tuberculosis Cysq, A Pap Phosphatase With Pap, Mg, And Li Bound
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2015-11-11 Classification: HYDROLASE Ligands: MG, LI, A3P |
 |
Structure Of M. Tuberculosis Cysq, A Pap Phosphatase With Amp, Po4, And 3Mg Bound
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2015-11-11 Classification: HYDROLASE Ligands: MG, AMP, PO4 |
 |
Structure Of M. Tuberculosis Cysq, A Pap Phosphatase With Amp, Po4, And 2Mg Bound
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2015-11-11 Classification: HYDROLASE Ligands: MG, AMP, PO4 |
 |
Structure Of M. Tuberculosis Cysq, A Pap Phosphatase With Po4 And 2Mg Bound
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2015-11-11 Classification: HYDROLASE Ligands: MG, NA, PO4 |
 |
Structure Of M. Tuberculosis Cysq, A Pap Phosphatase With Po4 And 2Ca Bound
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-11-11 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: CA, NA, PO4 |

