Search Count: 32
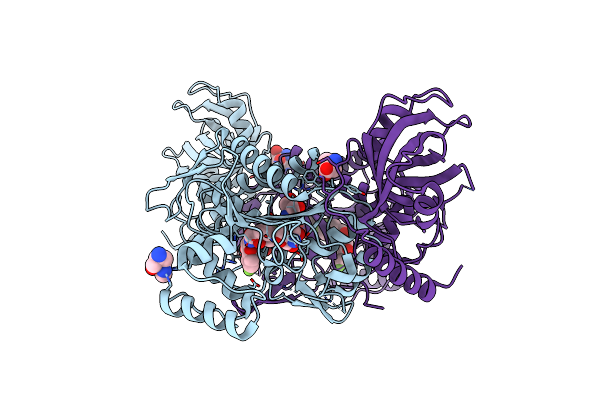 |
Crystal Structure Of Lysyl-Trna Synthetase From Plasmodium Falciparum Bound To A Difluoro Cyclohexyl Chromone Ligand
Organism: Plasmodium falciparum 3d7
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2019-04-03 Classification: TRANSFERASE Ligands: FYB, LYS, PEG, HIS, TRS |
 |
Crystal Structure Of Lysyl-Trna Synthetase From Plasmodium Falciparum Complexed With A Chromone Ligand
Organism: Plasmodium falciparum 3d7
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2019-04-03 Classification: TRANSFERASE Ligands: FYE |
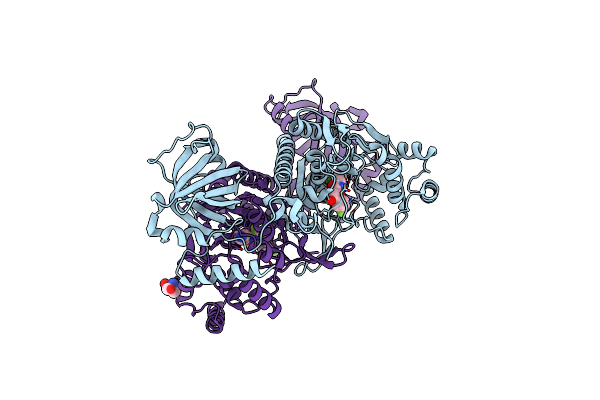 |
Crystal Structure Of Lysyl-Trna Synthetase From Cryptosporidium Parvum Complexed With L-Lysine And A Difluoro Cyclohexyl Chromone Ligand
Organism: Cryptosporidium parvum (strain iowa ii)
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2019-04-03 Classification: TRANSFERASE Ligands: LYS, FYB, TRS |
 |
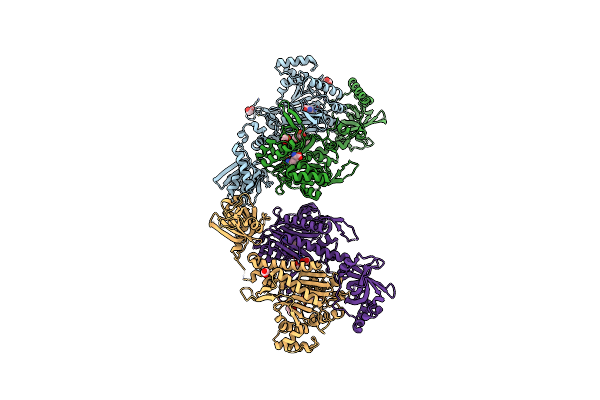
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2019-03-13 Classification: LIGASE/INHIBITOR Ligands: 9X0, CO, FMT, MLA, LYS |
 |
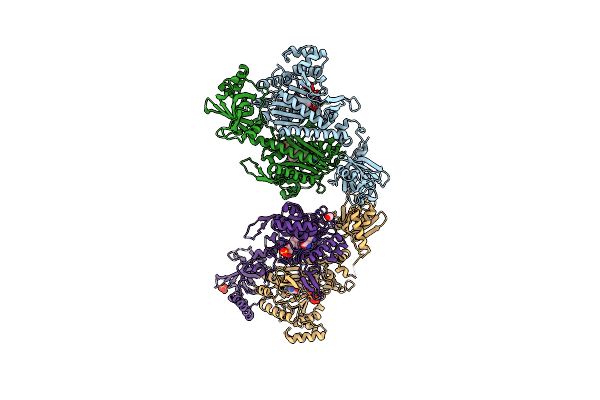
Crystal Structure Of Lysyl-Trna Synthetase From Cryptosporidium Parvum Complexed With L-Lysine
Organism: Cryptosporidium parvum (strain iowa ii)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-11-16 Classification: LIGASE Ligands: LYS, EDO, GOL |
 |
Crystal Structure Of Lysyl-Trna Synthetase From Cryptosporidium Parvum Complexed With L-Lysine And Cladosporin
Organism: Cryptosporidium parvum (strain iowa ii)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-11-16 Classification: ligase/ligase inhibitor Ligands: LYS, KRS, SO4, EDO |
 |
Organism: Dioclea grandiflora
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2015-07-22 Classification: SUGAR BINDING PROTEIN Ligands: CA, MN, SO4, GOL, XMM |
 |
 |
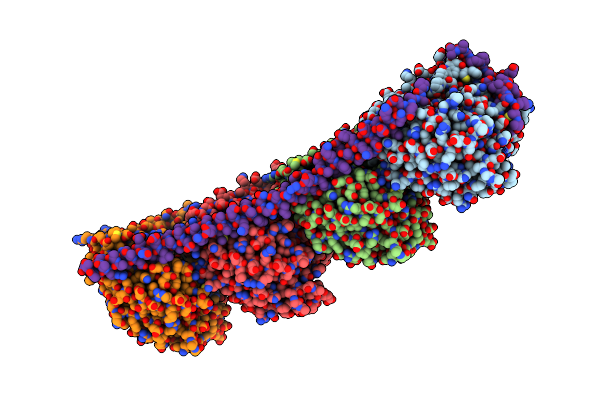 |
Organism: Rattus norvegicus, Ovis aries
Method: X-RAY DIFFRACTION Resolution:4.00 Å Release Date: 2010-07-28 Classification: CELL CYCLE Ligands: GTP, MG, GDP, G2N |
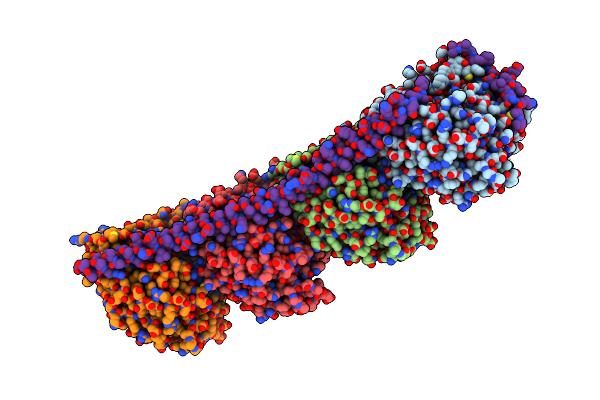 |
Organism: Rattus norvegicus, Ovis aries
Method: X-RAY DIFFRACTION Resolution:4.00 Å Release Date: 2010-07-28 Classification: CELL CYCLE Ligands: GTP, MG, GDP, K2N |
 |
 |
 |
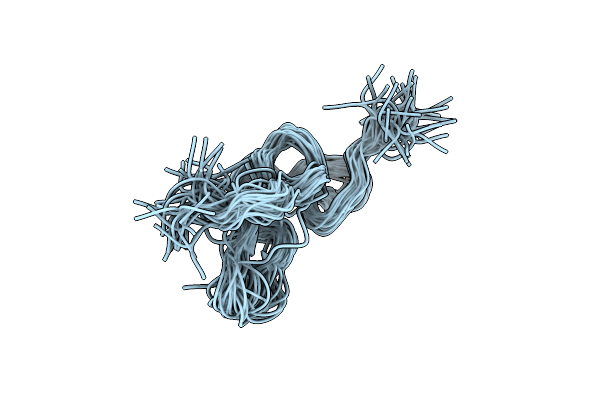
Solution Structure Of Jerdostatin From Trimeresurus Jerdonii With End C-Terminal Residues N45G46 Deleted
|
 |
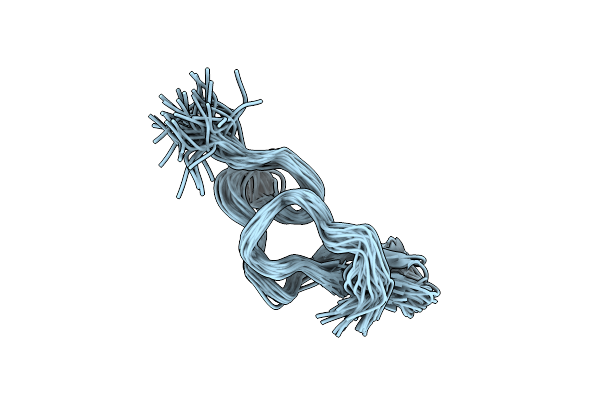
Solution Structure Of Jerdostatin Mutant R24K From Trimeresurus Jerdonii With End C-Terminal Residues N45G46 Deleted
|
 |
Organism: Roystonea regia
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2009-11-24 Classification: OXIDOREDUCTASE Ligands: HEM, CA, PEO, SO4, MES, EDO, NAG |
 |
Crystal Structure Of Recombinant Dioclea Guianensis Lectin Complexed With 5-Bromo-4-Chloro-3-Indolyl-A-D-Mannose
Organism: Dioclea guianensis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2007-10-30 Classification: CARBOHYDRATE-BINDING PROTEIN Ligands: MN, CA, CD, XMM |
 |
Crystal Structure Of Recombinant Dioclea Guianensis Lectin S131H Complexed With 5-Bromo-4-Chloro-3-Indolyl-A-D-Mannose
Organism: Dioclea guianensis
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2007-10-30 Classification: SUGAR BINDING PROTEIN Ligands: MN, CA, XMM |
 |
Crystal Structure Of Recombinant Dioclea Grandiflora Lectin Complexed With 5-Bromo-4-Chloro-3-Indolyl-A-D-Mannose
Organism: Dioclea grandiflora
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2007-10-30 Classification: SUGAR BINDING PROTEIN Ligands: CA, MN, XMM, SO4 |
 |
Crystal Structure Of Recombinant Dioclea Grandiflora Lectin Mutant E123A-H131N-K132Q Complexed With 5-Bromo-4-Chloro-3-Indolyl-A-D- Mannose
Organism: Dioclea grandiflora
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2007-10-30 Classification: SUGAR BINDING PROTEIN Ligands: CA, MN, XMM |

