Search Count: 35
 |
Organism: Mycolicibacterium chubuense, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: 6OU |
 |
Organism: Mycolicibacterium chubuense
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: A1A7X |
 |
Organism: Mycobacteroides abscessus
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: A1A8B, DSL |
 |
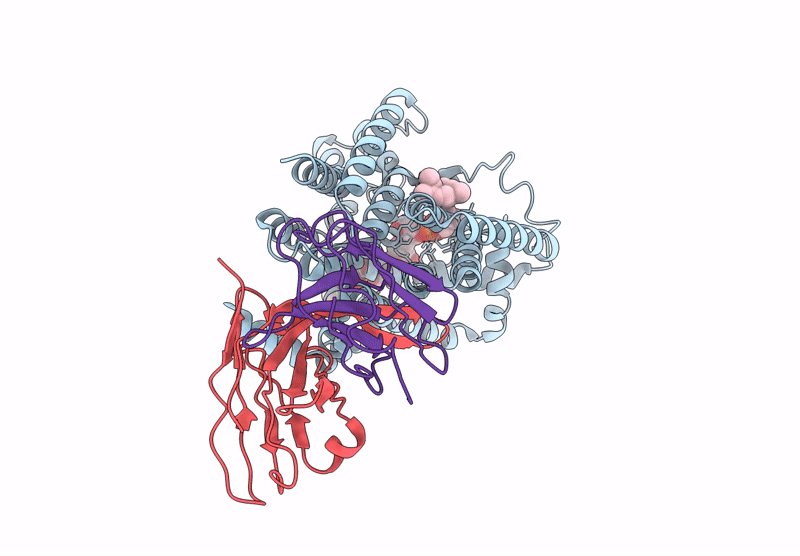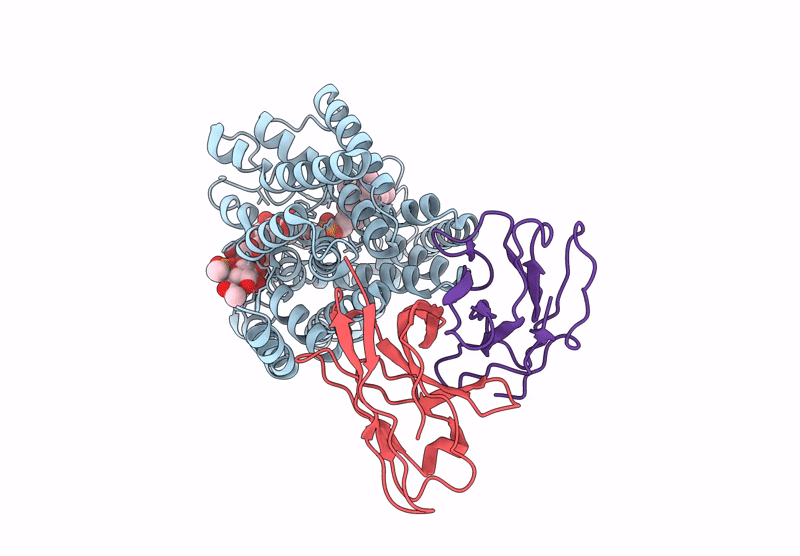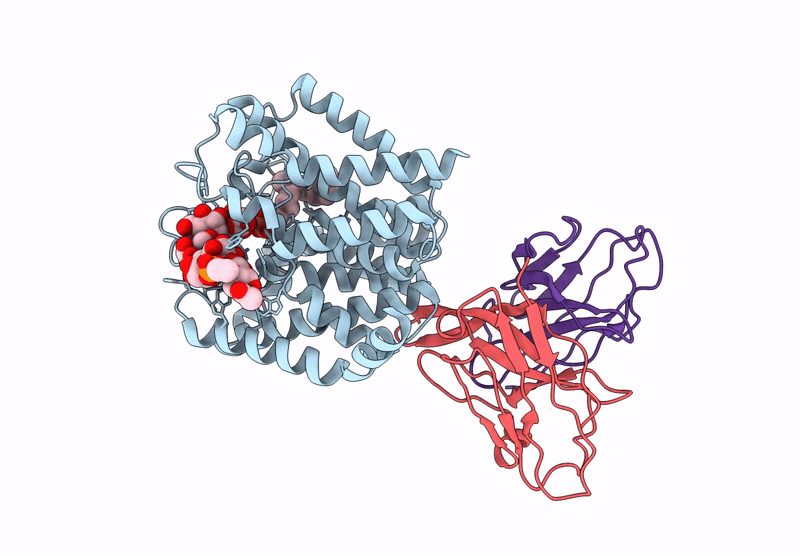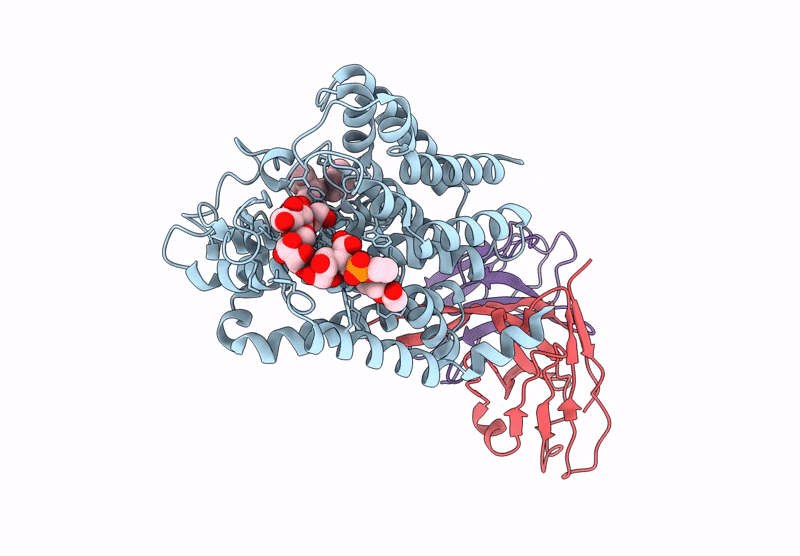
Organism: Mycobacteroides abscessus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN |
 |
Organism: Mycobacteroides abscessus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: A1A8B, DSL |
 |
Structure Of A Phosphatidylinositol-Phosphate Synthase (Pips) From Mycobacterium Kansasii
Organism: Archaeoglobus fulgidus, Mycobacterium kansasii
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2020-05-27 Classification: MEMBRANE PROTEIN Ligands: FLC, NA, PO4, TCE, OLC, 8K6, GOL, XP4 |
 |
Structure Of A Phosphatidylinositol-Phosphate Synthase (Pips) From Mycobacterium Kansasii With Evidence Of Substrate Binding
Organism: Archaeoglobus fulgidus, Mycobacterium kansasii atcc 12478
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2020-05-27 Classification: MEMBRANE PROTEIN Ligands: LIP, NA, TCE, 1PE, OLC, 8K6, GOL, C5P, FLC |
 |
High Resolution Semet Structure Of The Third Cohesin From Ruminococcus Flavefaciens Scaffoldin Protein, Scab
Organism: Ruminococcus flavefaciens
Method: X-RAY DIFFRACTION Resolution:1.14 Å Release Date: 2016-09-28 Classification: SUGAR BINDING PROTEIN Ligands: GOL |
 |
Structure Of A Phosphatidylinositolphosphate (Pip) Synthase From Renibacterium Salmoninarum
Organism: Archaeoglobus fulgidus, Renibacterium salmoninarum
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2015-11-11 Classification: MEMBRANE PROTEIN Ligands: SO4, 8K6, MG |
 |
Structure Of A Phosphatidylinositolphosphate (Pip) Synthase From Renibacterium Salmoninarum
Organism: Archaeoglobus fulgidus, Renibacterium salmoninarum
Method: X-RAY DIFFRACTION Resolution:3.62 Å Release Date: 2015-11-04 Classification: MEMBRANE PROTEIN Ligands: 8K6, MG, 58A |
 |
Crystal Structure Of Archaeoglobus Fulgidus Ipct-Dipps Bifunctional Membrane Protein
Organism: Archaeoglobus fulgidus
Method: X-RAY DIFFRACTION Resolution:2.66 Å Release Date: 2014-07-02 Classification: TRANSFERASE Ligands: LFA, MG |
 |
Structure Of An Atypical Alpha-Phosphoglucomutase Similar To Eukaryotic Phosphomannomutases
Organism: Lactococcus lactis subsp. cremoris mg1363
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2013-10-02 Classification: ISOMERASE Ligands: GOL, SO4 |
 |
Organism: Staphylococcus aureus
Method: SOLUTION NMR Release Date: 2012-09-12 Classification: HYDROLASE |
 |
The 3-Dimensional Structure Of Apo-Mpgp, The Mannosyl-3- Phosphoglycerate Phosphatase From Thermus Thermophilus Hb27 In Its Apo-Form
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2011-10-19 Classification: HYDROLASE Ligands: PO4 |
 |
The 3-Dimensional Structure Of The Gadolinium Derivative Of Mpgp, The Mannosyl-3-Phosphoglycerate Phosphatase From Thermus Thermophilus Hb27
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2011-10-19 Classification: HYDROLASE Ligands: GD, CL |
 |
The 3-Dimensional Structure Of Mpgp From Thermus Thermophilus Hb27, In Complex With The Alpha-Mannosylglycerate And Orthophosphate Reaction Products.
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2011-10-19 Classification: HYDROLASE Ligands: MG, PO4, 2M8 |
 |
The 3-Dimensional Structure Of Mpgp From Thermus Thermophilus Hb27, In Complex With The Alpha-Mannosylglycerate And Orthophosphate Reaction Products.
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2011-10-19 Classification: HYDROLASE Ligands: MG, PO4, 2M8 |
 |
The 3-Dimensional Structure Of Mpgp From Thermus Thermophilus Hb27, In Complex With The Alpha-Mannosylglycerate And Metaphosphate.
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2011-10-19 Classification: HYDROLASE Ligands: MG, PO4, CL, PO3, 2M8 |
 |
The 3-Dimensional Structure Of Mpgp From Thermus Thermophilus Hb27, In Complex With The Alpha-Mannosylglycerate.
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2011-10-19 Classification: HYDROLASE Ligands: 2M8, MG |
 |
The 3-Dimensional Structure Of Mpgp From Thermus Thermophilus Hb27, In Complex With The Metavanadate
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2011-10-19 Classification: HYDROLASE Ligands: MG, VN3 |

