Search Count: 114
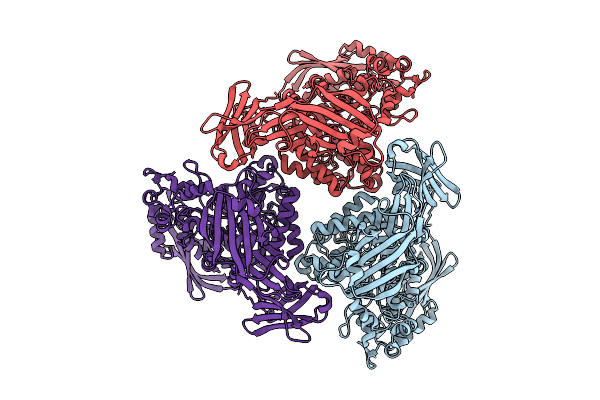 |
Active Conformation Of A Redox-Regulated Glycoside Hydrolase (Capgh2B) From The Gh2 Family
|
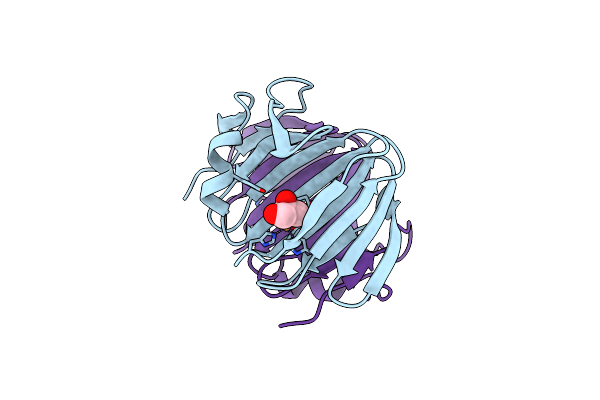 |
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-12-11 Classification: OXIDOREDUCTASE Ligands: GOL, CU |
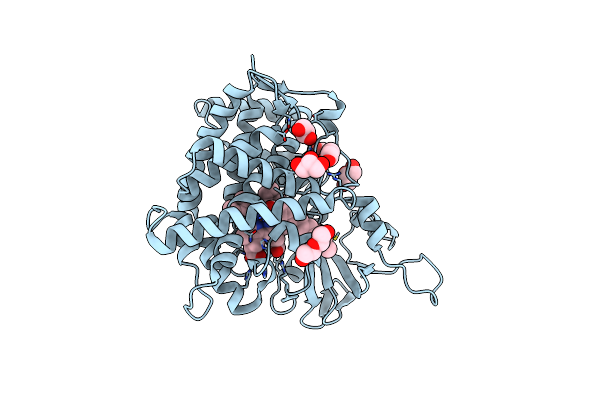 |
Crystal Structure Of A Fatty Acid Decarboxylase From Kocuria Marina In Complex With Myristic Acid
Organism: Kocuria marina
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2024-12-04 Classification: OXIDOREDUCTASE Ligands: MYR, HEM, GOL, PEG |
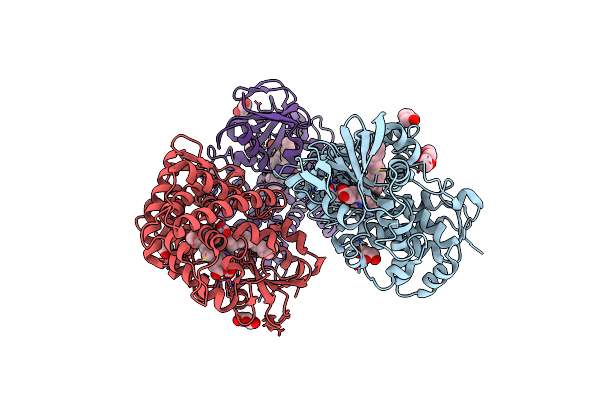 |
Crystal Structure Of A Fatty Acid Decarboxylase From Corynebacterium Lipophiloflavum In Complex With Palmitic Acid
Organism: Corynebacterium lipophiloflavum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-12-04 Classification: OXIDOREDUCTASE Ligands: CL, HEM, PLM, PEG, GOL, PGE |
 |
Crystal Structure Of A Fatty Acid Decarboxylase From Corynebacterium Lipophiloflavum In Complex With Oleic Acid
Organism: Corynebacterium lipophiloflavum
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-12-04 Classification: OXIDOREDUCTASE Ligands: HEM, OLA, PEG, GOL, CL, PGE |
 |
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-12-04 Classification: OXIDOREDUCTASE Ligands: CU |
 |
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2024-12-04 Classification: OXIDOREDUCTASE Ligands: GOL, CU |
 |
Organism: Bifidobacterium longum
Method: ELECTRON MICROSCOPY Release Date: 2022-11-16 Classification: HYDROLASE Ligands: ZN |
 |
Organism: Bifidobacterium longum
Method: ELECTRON MICROSCOPY Release Date: 2022-11-16 Classification: HYDROLASE Ligands: ZN |
 |
Organism: Bifidobacterium longum
Method: ELECTRON MICROSCOPY Release Date: 2022-11-16 Classification: HYDROLASE Ligands: ZN |
 |
Cryo-Em Structure Of Bl_Man38A Nucleophile Mutant In Complex With Mannose At 2.7 A
Organism: Bifidobacterium longum
Method: ELECTRON MICROSCOPY Release Date: 2022-11-16 Classification: HYDROLASE Ligands: ZN, MAN |
 |
Organism: Xanthomonas axonopodis pv. citri (strain 306)
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2020-12-02 Classification: HYDROLASE Ligands: CA, GOL, PEG |
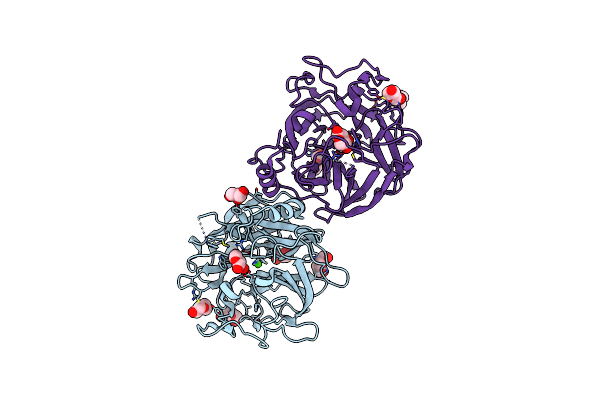 |
Crystal Structure Of The Gh43_1 Enzyme From Xanthomonas Citri Complexed With Xylose
Organism: Xanthomonas axonopodis pv. citri (strain 306)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-12-02 Classification: HYDROLASE Ligands: CA, XYP, PEG, GOL |
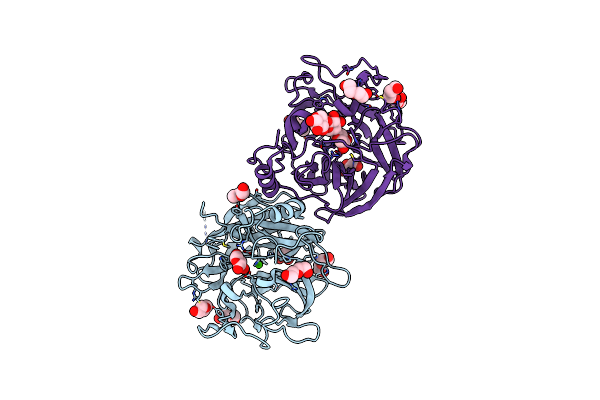 |
Crystal Structure Of The Gh43_1 Enzyme From Xanthomonas Citri Complexed With Xylotriose
Organism: Xanthomonas axonopodis pv. citri (strain 306)
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2020-12-02 Classification: HYDROLASE Ligands: CA, XYP, PEG, GOL |
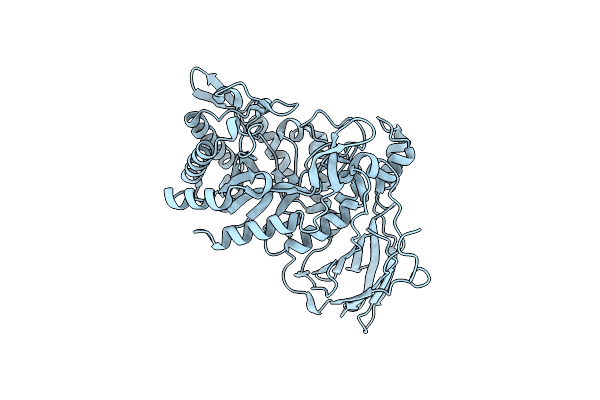 |
Organism: Xanthomonas axonopodis pv. citri (strain 306)
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2020-07-22 Classification: HYDROLASE |
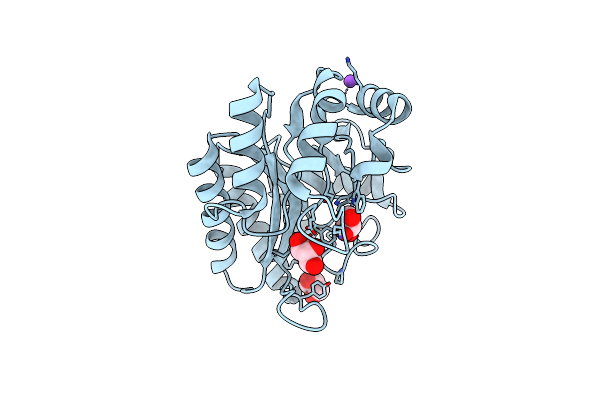 |
Crystal Structure Of A Gh128 (Subgroup I) Endo-Beta-1,3-Glucanase From Amycolatopsis Mediterranei (Amgh128_I)
Organism: Amycolatopsis mediterranei
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2020-05-20 Classification: HYDROLASE Ligands: GOL, NA |
 |
Crystal Structure Of A Gh128 (Subgroup I) Endo-Beta-1,3-Glucanase From Amycolatopsis Mediterranei (Amgh128_I) In Complex With Laminaritriose
Organism: Amycolatopsis mediterranei
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2020-05-20 Classification: HYDROLASE |
 |
Crystal Structure Of A Gh128 (Subgroup I) Endo-Beta-1,3-Glucanase (E199A Mutant) From Amycolatopsis Mediterranei (Amgh128_I) In Complex With Laminaripentaose
Organism: Amycolatopsis mediterranei
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2020-05-20 Classification: HYDROLASE Ligands: BGC, ZN, PEG |
 |
Crystal Structure Of A Gh128 (Subgroup I) Endo-Beta-1,3-Glucanase (E102A Mutant) From Amycolatopsis Mediterranei (Amgh128_I) In Complex With Laminaripentaose
Organism: Amycolatopsis mediterranei
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-05-20 Classification: HYDROLASE Ligands: ZN |
 |
Crystal Structure Of A Gh128 (Subgroup I) Endo-Beta-1,3-Glucanase (E102A Mutant) From Amycolatopsis Mediterranei (Amgh128_I) In Complex With Laminaritriose And Laminaribiose
Organism: Amycolatopsis mediterranei
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-05-20 Classification: HYDROLASE Ligands: ZN, PEG |

