Search Count: 15
 |
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-12-11 Classification: OXIDOREDUCTASE Ligands: GOL, CU |
 |
Crystal Structure Of A Fatty Acid Decarboxylase From Kocuria Marina In Complex With Myristic Acid
Organism: Kocuria marina
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2024-12-04 Classification: OXIDOREDUCTASE Ligands: MYR, HEM, GOL, PEG |
 |
Crystal Structure Of A Fatty Acid Decarboxylase From Corynebacterium Lipophiloflavum In Complex With Palmitic Acid
Organism: Corynebacterium lipophiloflavum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-12-04 Classification: OXIDOREDUCTASE Ligands: CL, HEM, PLM, PEG, GOL, PGE |
 |
Crystal Structure Of A Fatty Acid Decarboxylase From Corynebacterium Lipophiloflavum In Complex With Oleic Acid
Organism: Corynebacterium lipophiloflavum
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-12-04 Classification: OXIDOREDUCTASE Ligands: HEM, OLA, PEG, GOL, CL, PGE |
 |
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-12-04 Classification: OXIDOREDUCTASE Ligands: CU |
 |
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2024-12-04 Classification: OXIDOREDUCTASE Ligands: GOL, CU |
 |
Crystal Structure Of A Beta-Helix Domain Retrieved From Capybara Gut Metagenome
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-09-08 Classification: UNKNOWN FUNCTION Ligands: CA |
 |
Crystal Structure Of The Double Mutant L167W / P172L Of The Beta-Glucosidase From Trichoderma Harzianum
Organism: Trichoderma harzianum
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2019-06-26 Classification: HYDROLASE Ligands: NO3 |
 |
Organism: Xylella fastidiosa (strain 9a5c)
Method: X-RAY DIFFRACTION Resolution:2.63 Å Release Date: 2017-07-19 Classification: HYDROLASE Ligands: MN, IOD, PO4 |
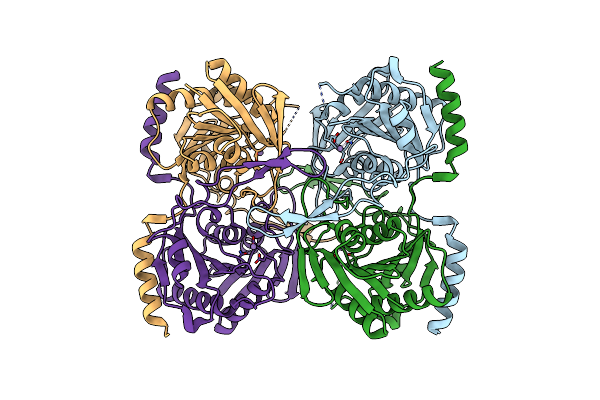 |
Stationary Phase Survival Protein E (Sure) From Xylella Fastidiosa - Xfsure-Tb (Tetramer Bigger).
Organism: Xylella fastidiosa (strain 9a5c)
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2017-07-19 Classification: HYDROLASE Ligands: MN, IOD, CL |
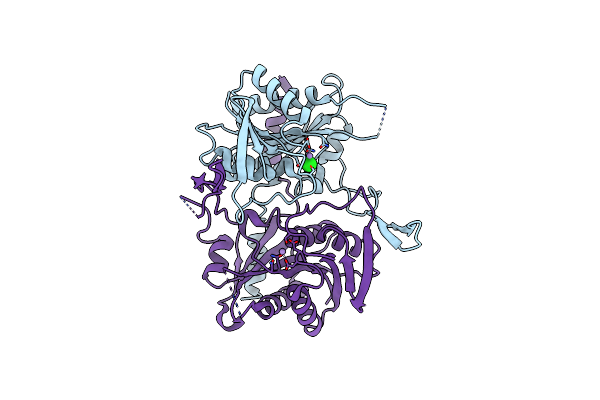 |
Stationary Phase Survival Protein E (Sure) From Xylella Fastidiosa - Xfsure-Ds (Dimer Smaller)
Organism: Xylella fastidiosa (strain 9a5c)
Method: X-RAY DIFFRACTION Resolution:2.82 Å Release Date: 2017-07-19 Classification: HYDROLASE Ligands: MN, IOD, CL |
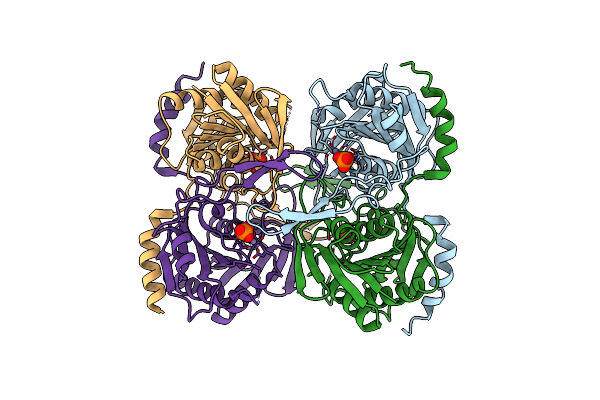 |
Stationary Phase Survival Protein E (Sure) From Xylella Fastidiosa- Xfsure-Tsamp (Tetramer Smaller - Crystallization With 3'Amp).
Organism: Xylella fastidiosa (strain 9a5c)
Method: X-RAY DIFFRACTION Resolution:2.76 Å Release Date: 2017-07-19 Classification: HYDROLASE Ligands: MN, IOD, PO4 |
 |
Crystal Structure Of A Small Heat-Shock Protein From Xylella Fastidiosa Reveals A Distinct High Order Structure
Organism: Xylella fastidiosa (strain 9a5c)
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2017-04-12 Classification: CHAPERONE |
 |
Organism: Trichoderma harzianum
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2016-07-06 Classification: HYDROLASE Ligands: GOL, SO4 |
 |
Organism: Xylella fastidiosa (strain 9a5c)
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2016-01-27 Classification: DNA BINDING PROTEIN |

