Search Count: 86
 |
Organism: Picea abies
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2025-03-12 Classification: OXIDOREDUCTASE Ligands: FAD, NAG, NA, GOL, CA |
 |
Crystal Structure Of The Catalytic Domain Of An Aa9 Lytic Polysaccharide Monooxygenase From Thermothelomyces Thermophilus (Ttlpmo9F)
Organism: Thermothelomyces thermophilus atcc 42464
Method: X-RAY DIFFRACTION Resolution:2.33 Å Release Date: 2024-12-25 Classification: METAL BINDING PROTEIN Ligands: PEG, PG4, EDO, FMT, CU |
 |
The Crystal Structure Of Rssymeg1 Reveals A Unique Form Of Smaller Gh7 Endoglucanases Alongside Gh7 Cellobiohydrolases In Protist Symbionts Of Termites
Organism: Reticulitermes speratus gut symbiotic protist
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2023-12-27 Classification: HYDROLASE Ligands: TRS, NA |
 |
Organism: Gloeophyllum trabeum atcc 11539
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2023-11-22 Classification: HYDROLASE |
 |
Organism: Phanerochaete chrysosporium
Method: X-RAY DIFFRACTION Resolution:2.54 Å Release Date: 2022-02-09 Classification: HYDROLASE Ligands: NAG, GOL |
 |
Organism: Phanerochaete chrysosporium
Method: X-RAY DIFFRACTION Resolution:3.08 Å Release Date: 2022-02-09 Classification: HYDROLASE Ligands: 1PE, PEG, EDO, NAG, XYP |
 |
Organism: Hypocrea jecorina (strain qm6a)
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2019-11-06 Classification: HYDROLASE Ligands: CO, NAG |
 |
Cellobiohydrolase I (Cel7A) From Trichoderma Reesei With S-Dihydroxypropranolol In The Active Site
Organism: Trichoderma reesei
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2018-10-10 Classification: HYDROLASE Ligands: NAG, CO, F9B |
 |
Organism: Heterobasidion irregulare tc 32-1
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2018-05-16 Classification: OXIDOREDUCTASE Ligands: NAG, CU, GOL, AKR, NA |
 |
Structural Studies Of A Glycoside Hydrolase Family 3 Beta-Glucosidase From The Model Fungus Neurospora Crassa
Organism: Neurospora crassa or74a
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2018-03-21 Classification: HYDROLASE Ligands: NAG |
 |
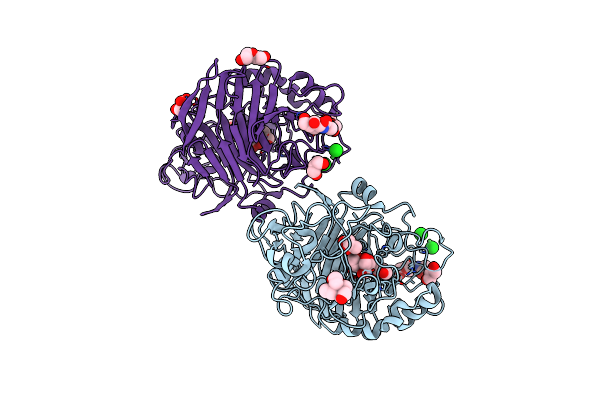
Organism: Hypocrea atroviridis
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2018-01-31 Classification: HYDROLASE Ligands: NAG, NI, GOL, BTB, GS1, GLC, PEG |
 |
Organism: Hypocrea atroviridis (strain atcc 20476 / imi 206040)
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2018-01-31 Classification: HYDROLASE Ligands: NAG, NI, BTB, GOL, CL, PEG |
 |
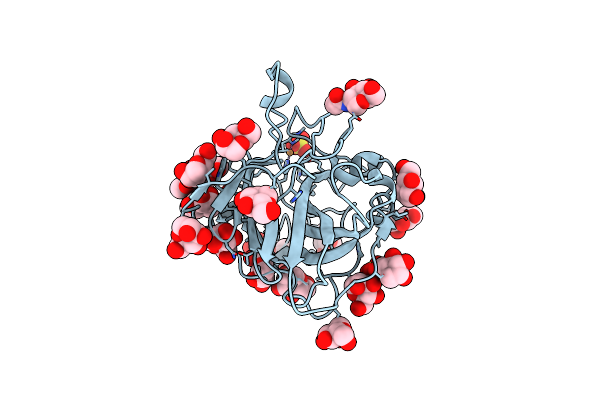
Organism: Trichoderma reesei qm6a
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-09-20 Classification: OXIDOREDUCTASE Ligands: NAG, MAN, CU, SO4 |
 |
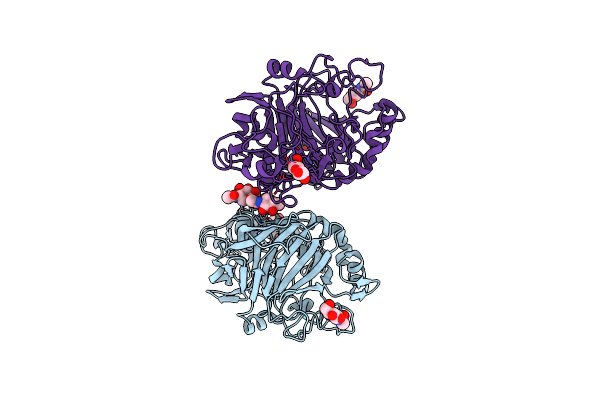
Organism: Trichoderma reesei qm6a
Method: X-RAY DIFFRACTION Resolution:0.95 Å Release Date: 2017-09-20 Classification: OXIDOREDUCTASE Ligands: NAG, MAN, CU, SO4 |
 |
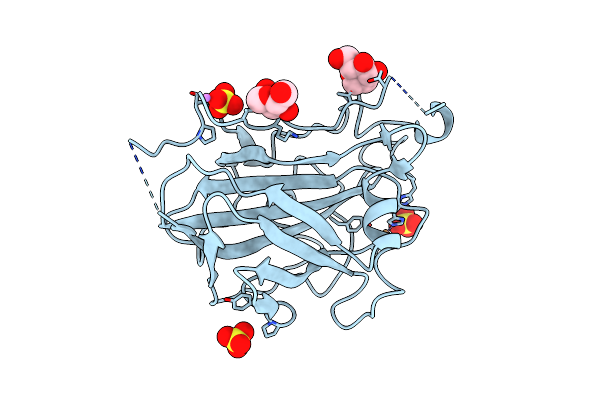
Cellobiohydrolase I (Cel7A) From Hypocrea Jecorina With Improved Thermal Stability
Organism: Hypocrea jecorina
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2017-09-06 Classification: HYDROLASE Ligands: NAG, GOL |
 |
Organism: Neurospora crassa
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2016-12-07 Classification: OXIDOREDUCTASE Ligands: MAN, CU, SO4, LI |
 |
The Structure Of Hypocrea Jecorina Beta-Xylosidase Xyl3A (Bxl1) In Complex With 4-Thioxylobiose
Organism: Hypocrea jecorina
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2016-09-21 Classification: HYDROLASE Ligands: NAG, ZN |
 |
Organism: Trichoderma reesei
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2016-08-10 Classification: HYDROLASE Ligands: NAG, ZN, TRS, GOL, ACT |
 |
Structural And Functional Studies Of Glycoside Hydrolase Family 3 Beta-Glucosidase Cel3A From The Moderately Thermophilic Fungus Rasamsonia Emersonii
Organism: Talaromyces emersonii
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2016-07-13 Classification: HYDROLASE Ligands: NAG, BGC, MAN |
 |
Organism: Tepidanaerobacter acetatoxydans
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2016-07-06 Classification: LIGASE Ligands: EDO, PEG, TAR, GOL, K, ACT |

