Search Count: 50
 |
Organism: Discosoma sp.
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: FLUORESCENT PROTEIN |
 |
Organism: Escherichia coli o157:h7, Branchiostoma lanceolatum
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2024-12-25 Classification: FLUORESCENT PROTEIN Ligands: ILE, GOL, CL |
 |
Organism: Escherichia coli, Escherichia coli o157:h7
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2024-12-25 Classification: FLUORESCENT PROTEIN Ligands: ILE, GOL, CL |
 |
Organism: Tick-borne encephalitis virus (strain sofjin)
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2024-10-09 Classification: VIRAL PROTEIN |
 |
Organism: Desulfurococcus amylolyticus
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2024-08-14 Classification: HYDROLASE |
 |
Inactivated Tick-Borne Encephalitis Virus (Tbev) Vaccine Strain Sofjin-Chumakov
Organism: Orthoflavivirus encephalitidis
Method: ELECTRON MICROSCOPY Release Date: 2024-06-05 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Tick-borne encephalitis virus (strain sofjin)
Method: ELECTRON MICROSCOPY Release Date: 2024-05-22 Classification: VIRUS Ligands: NAG |
 |
Organism: Cytaeis uchidae
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2023-12-27 Classification: FLUORESCENT PROTEIN Ligands: CL |
 |
Organism: Cytaeis uchidae
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-12-27 Classification: FLUORESCENT PROTEIN Ligands: CL |
 |
Organism: Cytaeis uchidae
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2023-12-27 Classification: FLUORESCENT PROTEIN Ligands: SO4, CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2023-07-05 Classification: OXIDOREDUCTASE Ligands: NAD, ZN, ACT |
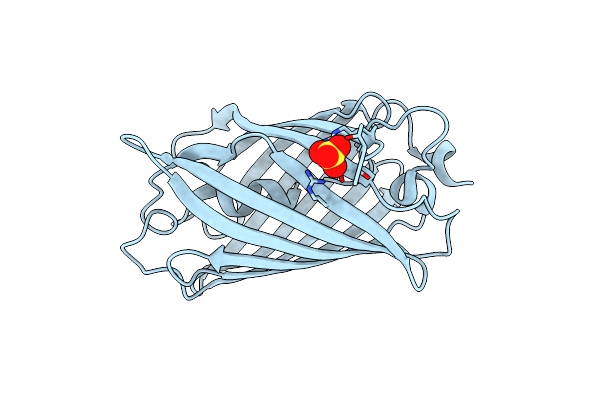 |
Organism: Discosoma sp.
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2022-11-02 Classification: FLUORESCENT PROTEIN Ligands: SO4 |
 |
Structure Of Beta-Lactamase Tem-171 Complexed With Tazobactam Intermediate At 2.3 A Resolution
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-07-13 Classification: HYDROLASE Ligands: TBE, ACT, TRS, EDO |
 |
Structure Of Beta-Lactamase Tem-171 Complexed With Tazobactam Intermediate At 2.5 A Resolution
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2022-07-13 Classification: HYDROLASE Ligands: TBE, ACT |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-07-13 Classification: HYDROLASE Ligands: EDO, TRS, ACT |
 |
Organism: Naja naja
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2022-04-27 Classification: TOXIN |
 |
Organism: Naja naja
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-04-27 Classification: TOXIN |
 |
Atomic Resolution X-Ray Structure Of The Uridine Phosphorylase From Vibrio Cholerae On Crystals Grown Under Microgravity
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:1.04 Å Release Date: 2021-06-30 Classification: TRANSFERASE Ligands: GOL, EDO, NA, PEG, MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2018-06-06 Classification: ISOMERASE Ligands: 6B9, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2018-06-06 Classification: ISOMERASE Ligands: SO4, 0FI |

