Search Count: 44
 |
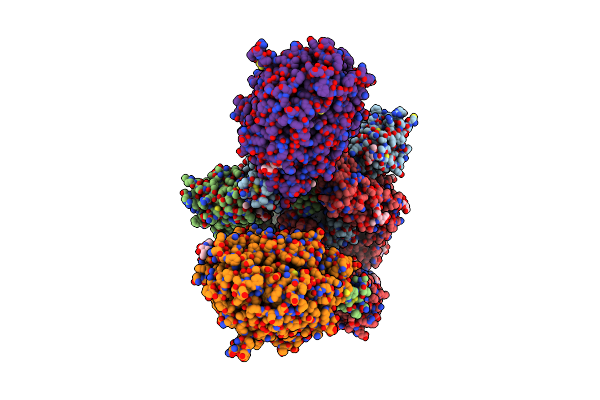
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-12-16 Classification: HYDROLASE/VIRAL PROTEIN Ligands: NAG |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-12-16 Classification: HYDROLASE/VIRAL PROTEIN Ligands: NAG |
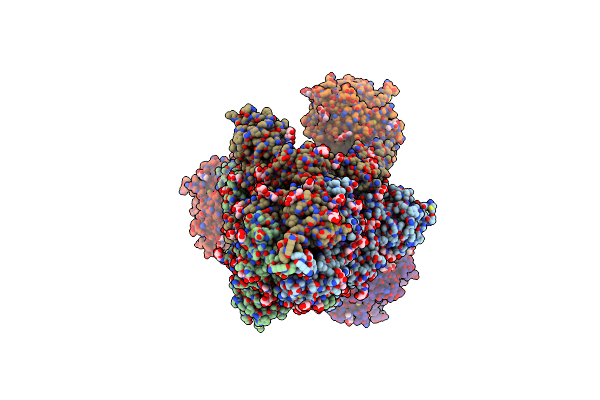 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-12-16 Classification: VIRAL PROTEIN/Hydrolase Ligands: NAG |
 |
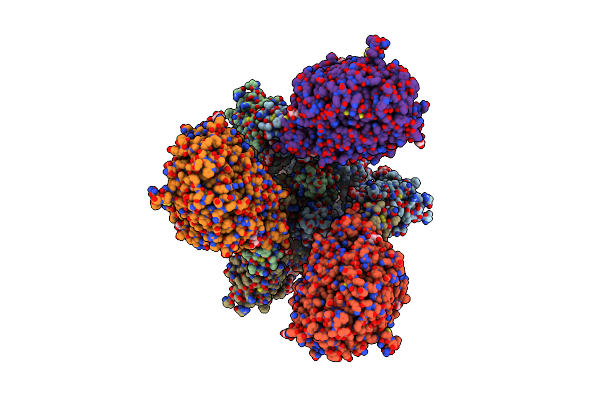
Ace2-Rbd Focused Refinement Using Symmetry Expansion Of Applied C3 For Triple Ace2-Bound Sars-Cov-2 Trimer Spike At Ph 7.4
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2020-12-09 Classification: Hydrolase/Viral Protein Ligands: NAG |
 |
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2020-12-09 Classification: HYDROLASE/VIRAL PROTEIN Ligands: NAG |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-12-09 Classification: HYDROLASE/VIRAL PROTEIN Ligands: NAG |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-12-09 Classification: HYDROLASE/VIRAL PROTEIN Ligands: NAG |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2020-11-25 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2020-08-12 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2020-08-12 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2020-08-12 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2020-08-12 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2020-07-29 Classification: VIRAL PROTEIN Ligands: NAG |
 |
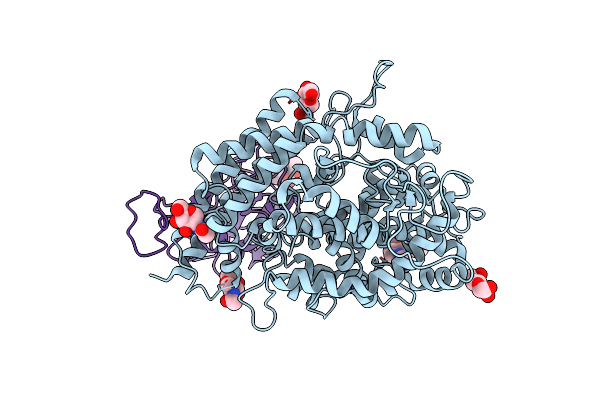
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:4.00 Å Release Date: 2016-10-19 Classification: MEMBRANE PROTEIN, TRANSPORT PROTEIN Ligands: NAG |
 |
Ampa Subtype Ionotropic Glutamate Receptor Glua2 In Complex With Noncompetitive Inhibitor Cp465022
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:4.37 Å Release Date: 2016-10-19 Classification: TRANSPORT PROTEIN/INHIBITOR Ligands: NAG, 6ZQ |
 |
Ampa Subtype Ionotropic Glutamate Receptor Glua2 In Complex With Noncompetitive Inhibitor Perampanel
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:4.00 Å Release Date: 2016-10-19 Classification: TRANSPORT PROTEIN/INHIBITOR Ligands: NAG, 6ZP |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:4.51 Å Release Date: 2016-10-19 Classification: TRANSPORT PROTEIN/INHIBITOR Ligands: NAG, GYB |
 |
Ampa Subtype Ionotropic Glutamate Receptor Glua2 In Complex With Noncompetitive Inhibitor Gyki53655
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:3.80 Å Release Date: 2016-10-19 Classification: TRANSPORT PROTEIN/INHIBITOR Ligands: NAG, GYK |
 |
Crystal Structure Of Repressor Of Toxin (Rot), A Central Regulator Of Staphylococcus Aureus Virulence
Organism: Staphylococcus aureus subsp. aureus cn1
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2014-11-05 Classification: DNA BINDING PROTEIN Ligands: CL |
 |
Crystal Structure Of Rabbit Mab R56 Fab In Complex With V3 Crown Of Hiv-1 Jr-Fl Gp120
Organism: Oryctolagus cuniculus, Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2013-07-31 Classification: IMMUNE SYSTEM/VIRAL PROTEIN Ligands: CA |

