Search Count: 18
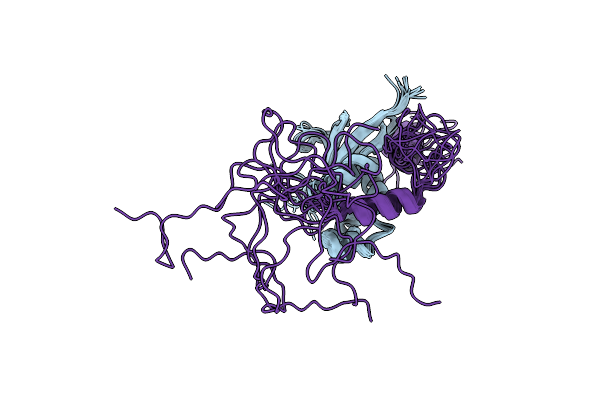 |
Structure Of Sars-Cov-2 Nucleoprotein In Dynamic Complex With Its Viral Partner Nsp3A
Organism: Severe acute respiratory syndrome coronavirus 2
Method: SOLUTION NMR Release Date: 2022-01-19 Classification: VIRAL PROTEIN |
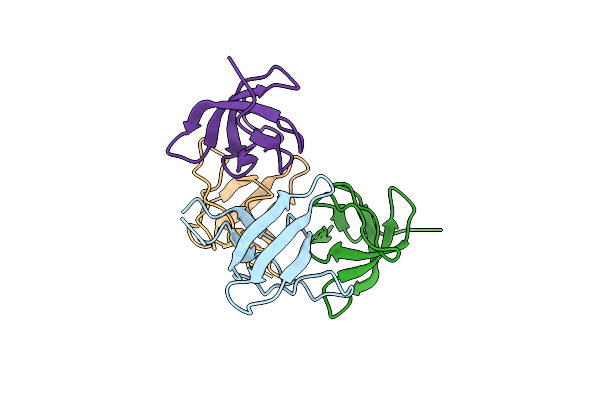 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2021-12-22 Classification: SIGNALING PROTEIN |
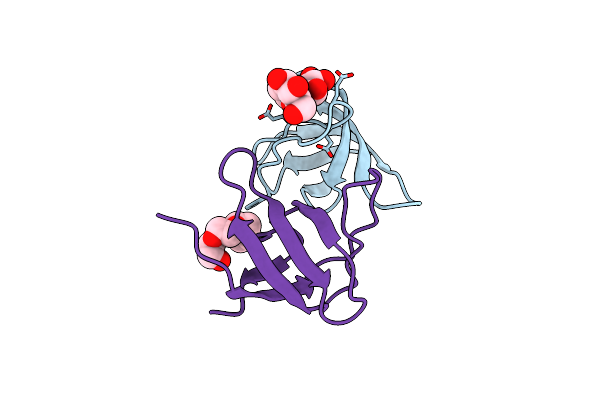 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2021-12-22 Classification: SIGNALING PROTEIN Ligands: PG4 |
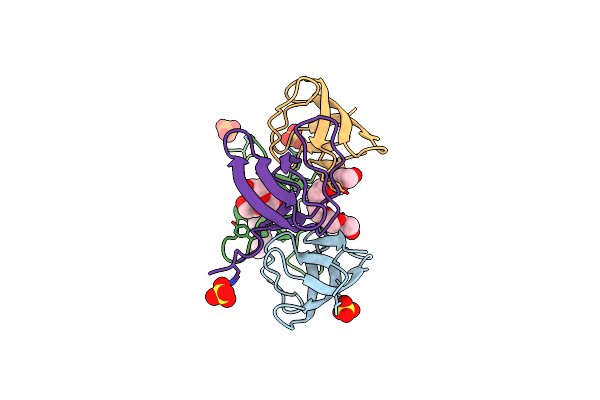 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2021-12-22 Classification: SIGNALING PROTEIN Ligands: SO4, P6G, PO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2021-12-22 Classification: SIGNALING PROTEIN Ligands: PO4, 1PE, EDO, PG4, SO4, PEG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2021-12-22 Classification: SIGNALING PROTEIN Ligands: PG4, SO4, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2021-12-22 Classification: SIGNALING PROTEIN Ligands: PO4, PG4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.11 Å Release Date: 2021-12-22 Classification: SIGNALING PROTEIN Ligands: EPE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2021-12-22 Classification: SIGNALING PROTEIN Ligands: EDO, PEG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2016-08-10 Classification: IMMUNE SYSTEM |
 |
The Crystal Structure Of Beta2-Microglobulin D76N Mutant At Room Temperature
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2016-08-10 Classification: IMMUNE SYSTEM |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2016-08-10 Classification: IMMUNE SYSTEM Ligands: ACT |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2015-11-18 Classification: IMMUNE SYSTEM Ligands: PGE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2015-11-18 Classification: IMMUNE SYSTEM Ligands: ACT, PGE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2015-11-18 Classification: IMMUNE SYSTEM Ligands: PGE, ACT |
 |
The Co-Complex Structure Of The Translation Initiation Factor Eif4E With The Inhibitor 4Egi-1 Reveals An Allosteric Mechanism For Dissociating Eif4G
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2014-08-13 Classification: PROTEIN BINDING Ligands: MGP, 33R, MES |
 |
The Co-Complex Structure Of The Translation Initiation Factor Eif4E With The Inhibitor 4Egi-1 Reveals An Allosteric Mechanism For Dissociating Eif4G
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2014-08-13 Classification: PROTEIN BINDING Ligands: MGT, 34K, MES |
 |
The Co-Complex Structure Of The Translation Initiation Factor Eif4E With The Inhibitor 4Egi-1 Reveals An Allosteric Mechanism For Dissociating Eif4G
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-08-13 Classification: PROTEIN BINDING Ligands: M7G, 34J |

