Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
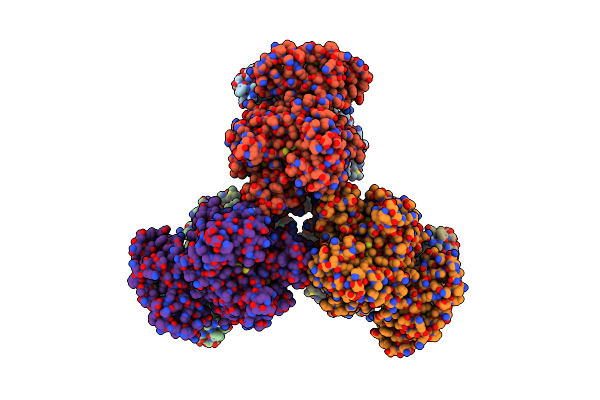 |
Cryoem Structure Of The Salmonella Effector Inositol Phosphate Phosphatase Sopb
Organism: Salmonella typhimurium (strain lt2 / sgsc1412 / atcc 700720)
Method: ELECTRON MICROSCOPY Release Date: 2024-07-10 Classification: LIPID BINDING PROTEIN |
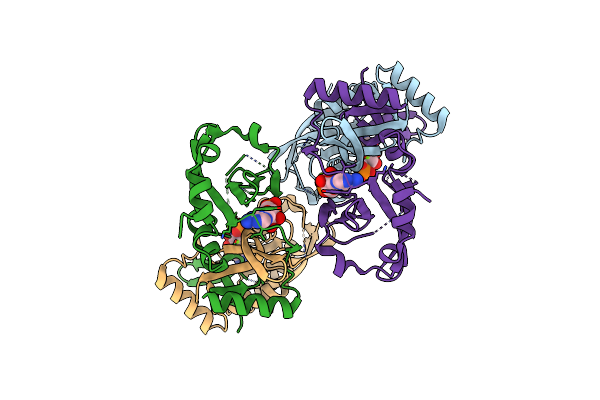 |
Organism: Salmonella typhimurium (strain 14028s / sgsc 2262)
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2024-02-28 Classification: TRANSFERASE Ligands: AMP, MG |
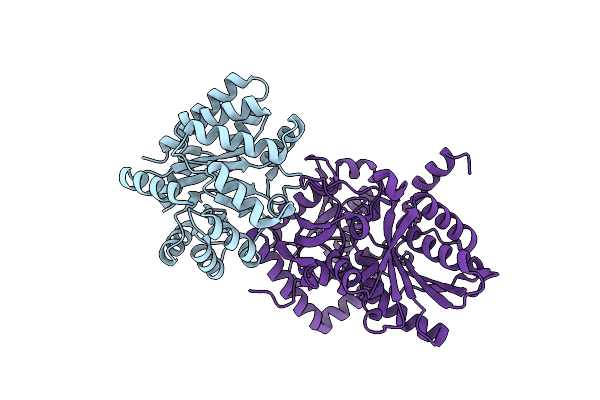 |
X-Ray Crystal Structure Of Salmonella Typhimurium Tryptophan Synthase Internal Aldimine At Ph 5.0
Organism: Salmonella typhimurium (strain lt2 / sgsc1412 / atcc 700720)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-02-14 Classification: LYASE |
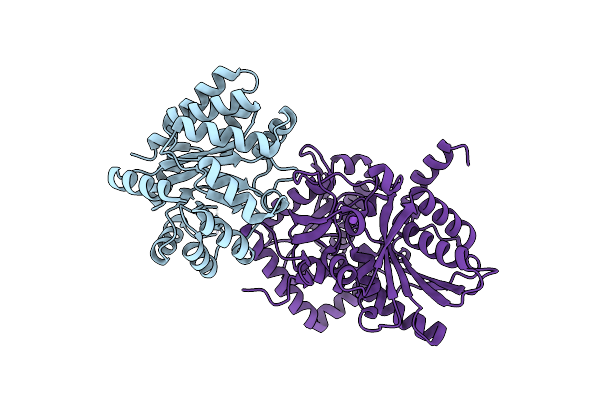 |
X-Ray Crystal Structure Of Salmonella Typhimurium Tryptophan Synthase Internal Aldimine
Organism: Salmonella typhimurium (strain lt2 / sgsc1412 / atcc 700720)
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-02-14 Classification: LYASE |
 |
Crystal Structure Of A68P Single Mutant Of O-Acetylserine Sulfhydrylase From Haemophilus Influenzae In Complex With High-Affinity Inhibitory Peptide From Serine Acetyltransferase Of Salmonella Typhimurium At 2.1 A
Organism: Haemophilus influenzae rd kw20, Salmonella typhimurium
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-08-02 Classification: TRANSFERASE/TRANSFERASE INHIBITOR |
 |
Crystal Structure Of Tetra Mutant (D67E,A68P,L98I,A301S) Of O-Acetylserine Sulfhydrylase From Salmonella Typhimurium In Complex With High-Affinity Inhibitory Peptide From Serine Acetyltransferase Of Salmonella Typhimurium At 2.8 A
Organism: Haemophilus influenzae rd kw20, Salmonella typhimurium
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2023-08-02 Classification: TRANSFERASE/TRANSFERASE INHIBITOR |
 |
Crystal Structure Of The Albicidin Resistance Protein Stm3175 From Salmonella Typhimurium
Organism: Salmonella typhimurium
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2022-09-28 Classification: DNA BINDING PROTEIN |
 |
Ruvab Branch Migration Motor Complexed To The Holliday Junction - Ruvb Aaa+ State S1 [T2 Dataset]
Organism: Streptococcus thermophilus, Salmonella typhimurium, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-09-14 Classification: HYDROLASE Ligands: AGS, MG, ADP |
 |
Ruvab Branch Migration Motor Complexed To The Holliday Junction - Ruvb Aaa+ State S2 [T2 Dataset]
Organism: Streptococcus thermophilus, Salmonella typhimurium, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-09-14 Classification: HYDROLASE Ligands: AGS, MG, ADP |
 |
Ruvab Branch Migration Motor Complexed To The Holliday Junction - Ruvb Aaa+ State S3 [T2 Dataset]
Organism: Streptococcus thermophilus, Salmonella typhimurium, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-09-14 Classification: HYDROLASE Ligands: ADP, MG, AGS |
 |
Ruvab Branch Migration Motor Complexed To The Holliday Junction - Ruvb Aaa+ State S4 [T2 Dataset]
Organism: Streptococcus thermophilus, Salmonella typhimurium, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-09-14 Classification: HYDROLASE Ligands: ADP, AGS, MG |
 |
Ruvab Branch Migration Motor Complexed To The Holliday Junction - Ruvb Aaa+ State S5 [T2 Dataset]
Organism: Streptococcus thermophilus, Salmonella typhimurium, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-09-14 Classification: HYDROLASE Ligands: ADP, AGS, MG |
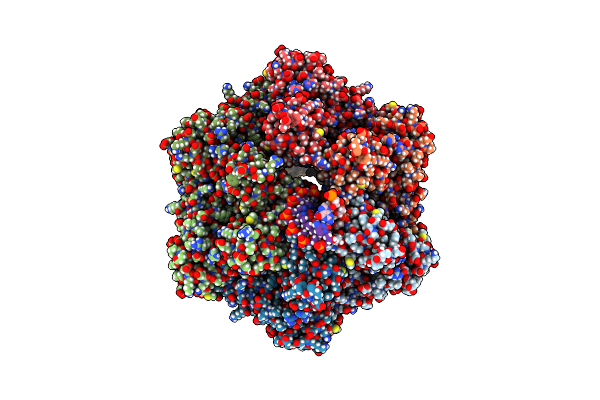 |
Ruvab Branch Migration Motor Complexed To The Holliday Junction - Ruvb Aaa+ State S0+A [T2 Dataset]
Organism: Streptococcus thermophilus, Salmonella typhimurium, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-09-14 Classification: HYDROLASE Ligands: AGS, MG, ADP |
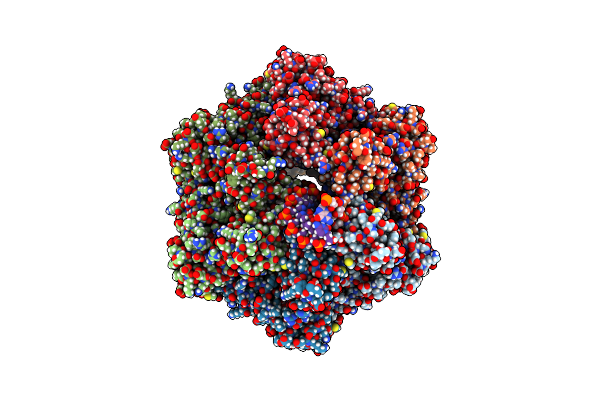 |
Ruvab Branch Migration Motor Complexed To The Holliday Junction - Ruvb Aaa+ State S0+A [T1 Dataset]
Organism: Streptococcus thermophilus, Salmonella typhimurium, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-09-14 Classification: HYDROLASE Ligands: AGS, MG, ADP |
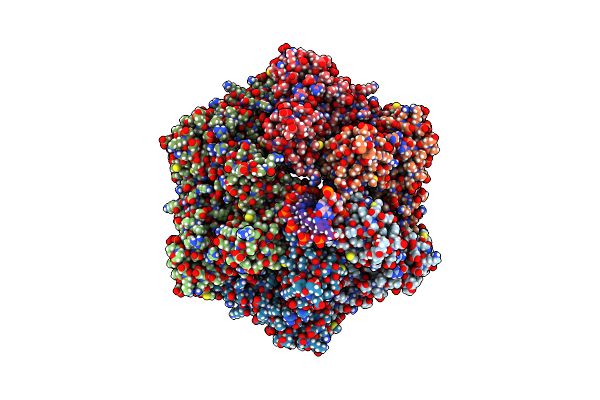 |
Ruvab Branch Migration Motor Complexed To The Holliday Junction - Ruvb Aaa+ State S1 [T1 Dataset]
Organism: Streptococcus thermophilus, Salmonella typhimurium, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-09-14 Classification: HYDROLASE Ligands: AGS, MG, ADP |
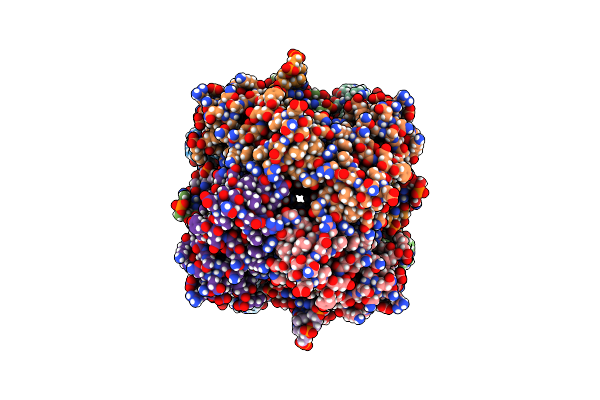 |
Ruvab Branch Migration Motor Complexed To The Holliday Junction - Ruva-Hj Core [T2 Dataset]
Organism: Salmonella typhimurium, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-09-14 Classification: HYDROLASE |
 |
Organism: Salmonella typhimurium
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2022-09-14 Classification: TOXIN/ANTITOXIN |
 |
Organism: Salmonella typhimurium (strain lt2 / sgsc1412 / atcc 700720)
Method: X-RAY DIFFRACTION Resolution:2.87 Å Release Date: 2022-05-04 Classification: DNA BINDING PROTEIN Ligands: HOH |
 |
The Internal Aldimine Form Of The Wild-Type Salmonella Typhimurium Tryptophan Synthase In Complex With Two Molecules Of N-(4'-Trifluoromethoxybenzoyl)-2-Amino-1-Ethylphosphate (F6F) Inhibitor At The Enzyme Alpha-Site, A Single F6F Molecule At The Enzyme Beta-Site, And Sodium Ion At The Metal Coordination Site At 1.55 Angstrom Resolution. One Of The Beta-Q114 Rotamer Conformations Allows A Hydrogen Bond To Form With The Plp Oxygen At The Position 3 In The Ring
Organism: Salmonella typhimurium (strain lt2 / sgsc1412 / atcc 700720)
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2022-03-09 Classification: LYASE/LYASE INHIBITOR Ligands: F6F, CL, PLP, PEG, EDO, NA |
 |
The Internal Aldimine Form Of The Wild-Type Salmonella Typhimurium Tryptophan Synthase In Complex With Cesium Ion At The Metal Coordination Site And The Product L-Tryptophan At The Enzyme Beta-Site At 1.60 Angstrom Resolution
Organism: Salmonella typhimurium (strain lt2 / sgsc1412 / atcc 700720)
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-03-02 Classification: LYASE Ligands: DMS, EDO, CS, CL, PLP, PEG, TRP |