Search Count: 59
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.56 Å Release Date: 2024-05-15 Classification: DNA BINDING PROTEIN |
 |
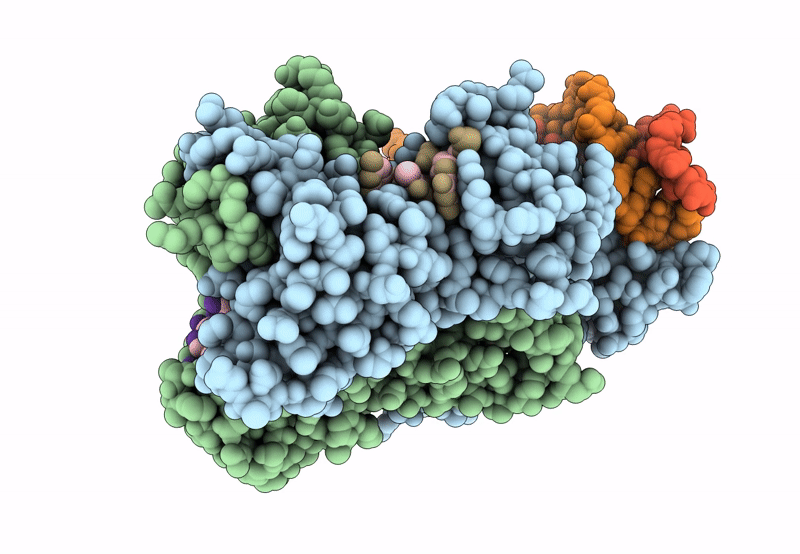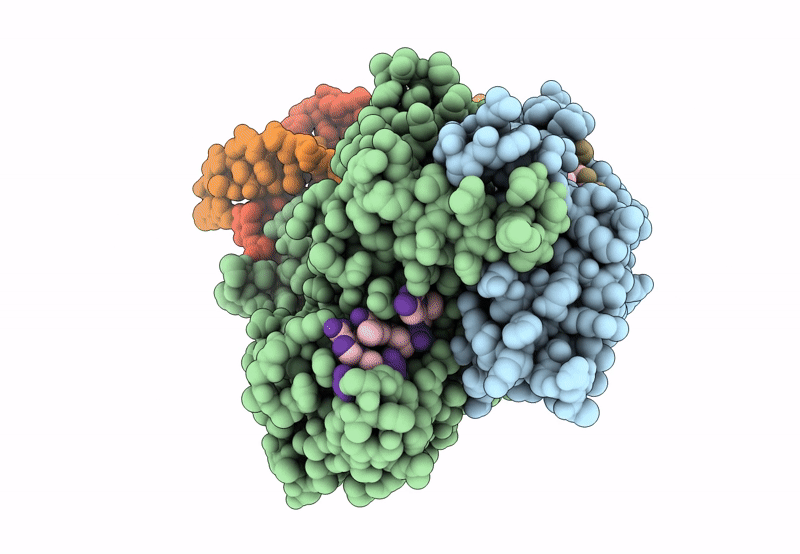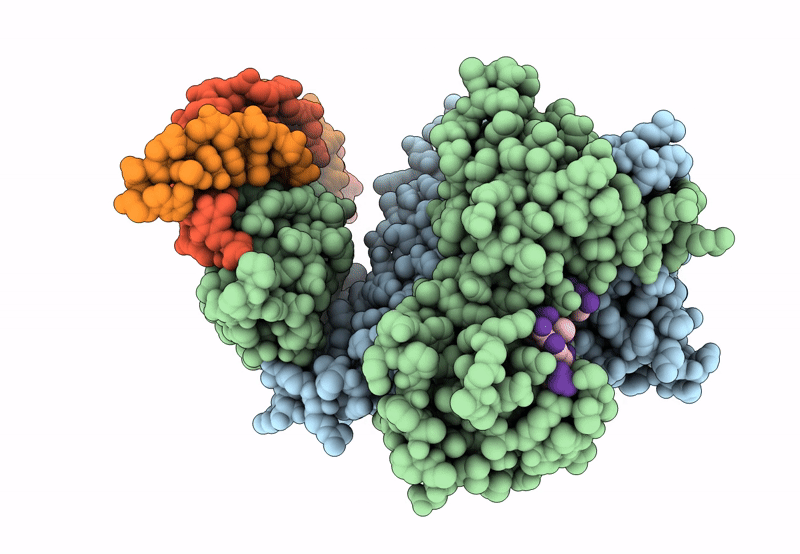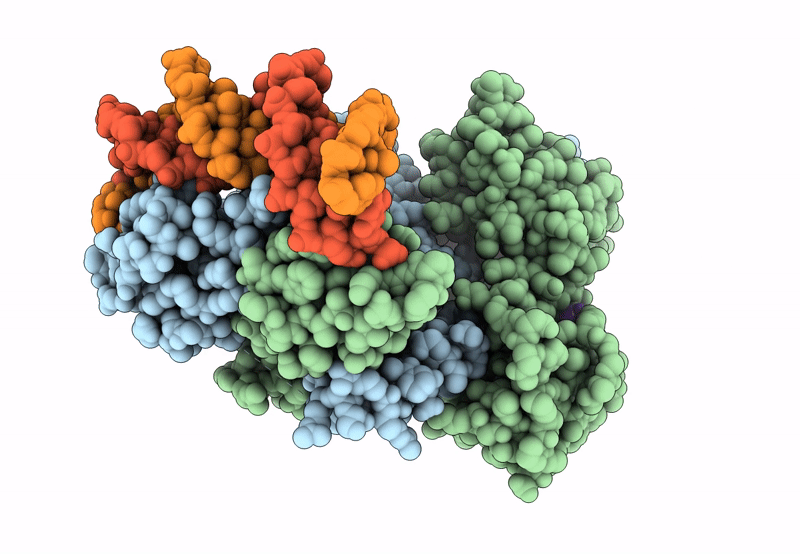
Structure Of The C. Crescentus Wyl-Activator, Drid, Bound To Ssdna And Cognate Dna
Organism: Caulobacter vibrioides na1000, Caulobacter vibrioides
Method: X-RAY DIFFRACTION Resolution:2.74 Å Release Date: 2023-11-29 Classification: TRANSCRIPTION/DNA Ligands: SO4 |
 |
Organism: Caulobacter vibrioides na1000, Caulobacter vibrioides, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.46 Å Release Date: 2023-11-29 Classification: TRANSCRIPTION/DNA |
 |
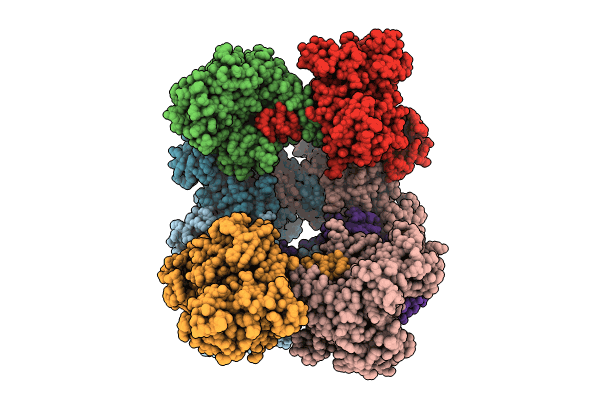
Cryo-Em Structure Of The Methanosarcina Mazei Apo Glutamin Synthetase Structure: Dodecameric Form
Organism: Methanosarcina mazei
Method: ELECTRON MICROSCOPY Release Date: 2023-11-15 Classification: LIGASE Ligands: MG |
 |
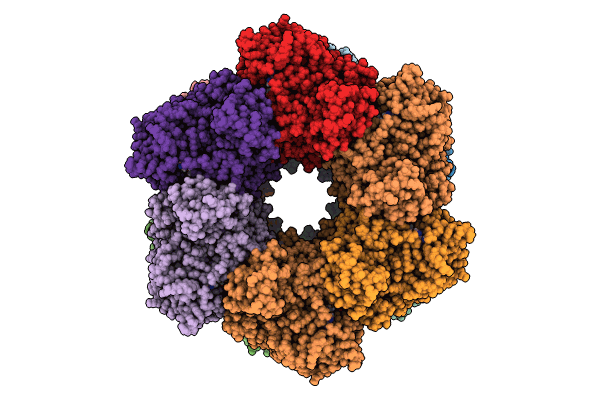
Cryo-Em Structure Of Methanosarcina Mazie Glutamine Synthetase Captured As Partial Oligomer
Organism: Methanosarcina mazei go1
Method: ELECTRON MICROSCOPY Release Date: 2023-11-15 Classification: LIGASE |
 |
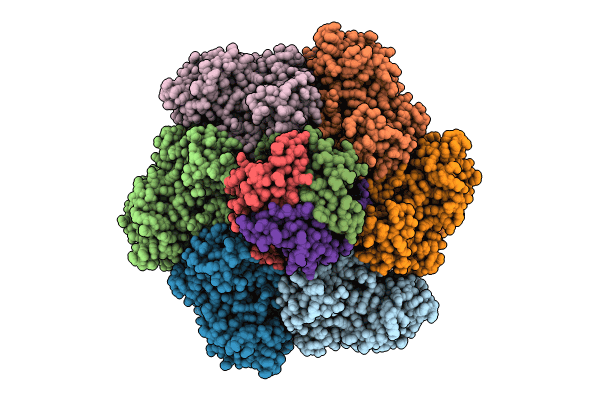
Cryo-Em Structure Of The Methanosarcina Mazei Glutamine Synthetase (Gs) With Met-Sox-P And Adp
Organism: Methanosarcina mazei go1
Method: ELECTRON MICROSCOPY Release Date: 2023-11-15 Classification: LIGASE Ligands: MG, P3S, ADP |
 |
Crystal Structure Of The Methanosarcina Mazei Glutamine Synthetase In Complex With Glnk1
Organism: Methanosarcina mazei go1
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-11-15 Classification: LYASE |
 |
Organism: Methanosarcina mazei go1
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2023-11-15 Classification: LIGASE Ligands: MG |
 |
Organism: Mycobacteriaceae
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2023-11-01 Classification: TRANSCRIPTION Ligands: YT9 |
 |
Organism: Rhodococcus sp. usk13, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2023-11-01 Classification: TRANSCRIPTION Ligands: SO4 |
 |
Organism: Mycolicibacterium smegmatis (mycobacterium smegmatis)
Method: X-RAY DIFFRACTION Resolution:3.56 Å Release Date: 2023-11-01 Classification: TRANSCRIPTION |
 |
Organism: Rhodococcus sp. usk13, Rhodococcus
Method: X-RAY DIFFRACTION Resolution:2.96 Å Release Date: 2023-11-01 Classification: TRANSCRIPTION/DNA Ligands: PO4 |
 |
Structure Of M. Baixiangningiae Darr-Dna Complex Reveals Novel Dimer-Of-Dimers Dna Binding
Organism: Mycolicibacterium baixiangningiae
Method: X-RAY DIFFRACTION Resolution:3.49 Å Release Date: 2023-11-01 Classification: TRANSCRIPTION/DNA |
 |
Organism: Rhodococcus sp. usk13
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2023-11-01 Classification: TRANSCRIPTION Ligands: CMP |
 |
Organism: Trypanosoma brucei
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2023-02-22 Classification: PROTEIN BINDING |
 |
Structure Of The Apo Form Of Streptomyces Venezuelae Glgx, The Glycogen Debranching Enzyme
Organism: Streptomyces venezuelae
Method: X-RAY DIFFRACTION Resolution:3.51 Å Release Date: 2022-10-05 Classification: HYDROLASE |
 |
Organism: Streptomyces venezuelae
Method: X-RAY DIFFRACTION Resolution:3.34 Å Release Date: 2022-10-05 Classification: HYDROLASE Ligands: C2E |
 |
Organism: Streptomyces venezuelae
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2022-10-05 Classification: HYDROLASE Ligands: A16, C2E, PO4 |
 |
Organism: Streptomyces venezuelae
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2022-10-05 Classification: HYDROLASE Ligands: C2E |
 |
Structure Of The Dna Binding Domain Of Psk1 Par Partition Protein Bound To Centromere Dna
Organism: Unidentified plasmid, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2022-08-10 Classification: DNA BINDING PROTEIN/DNA |

