Search Count: 29
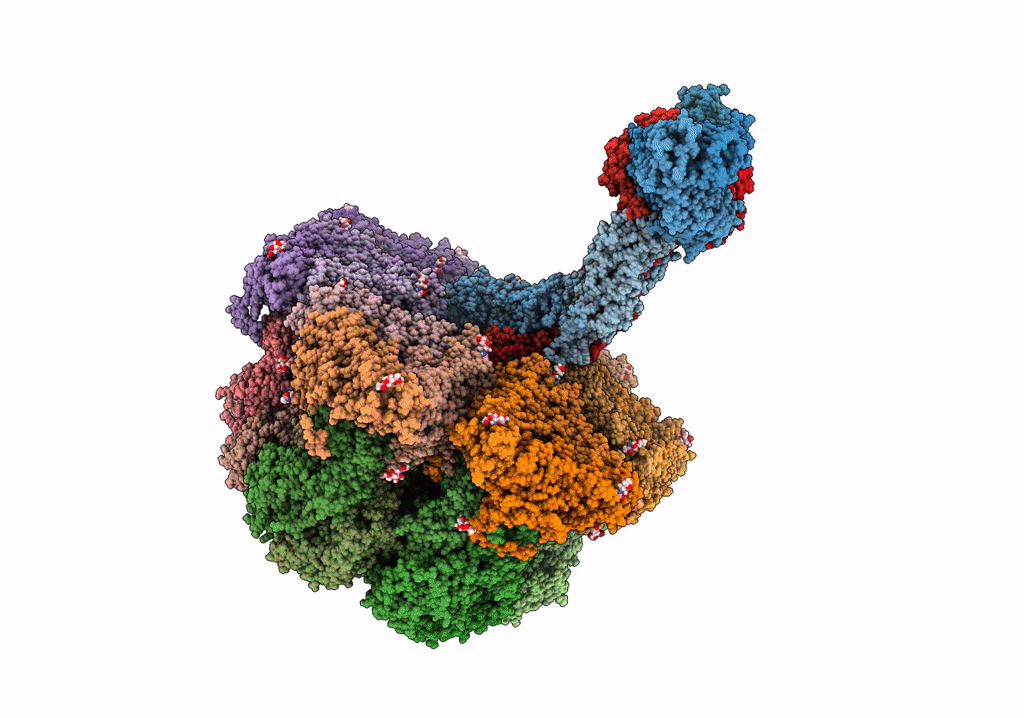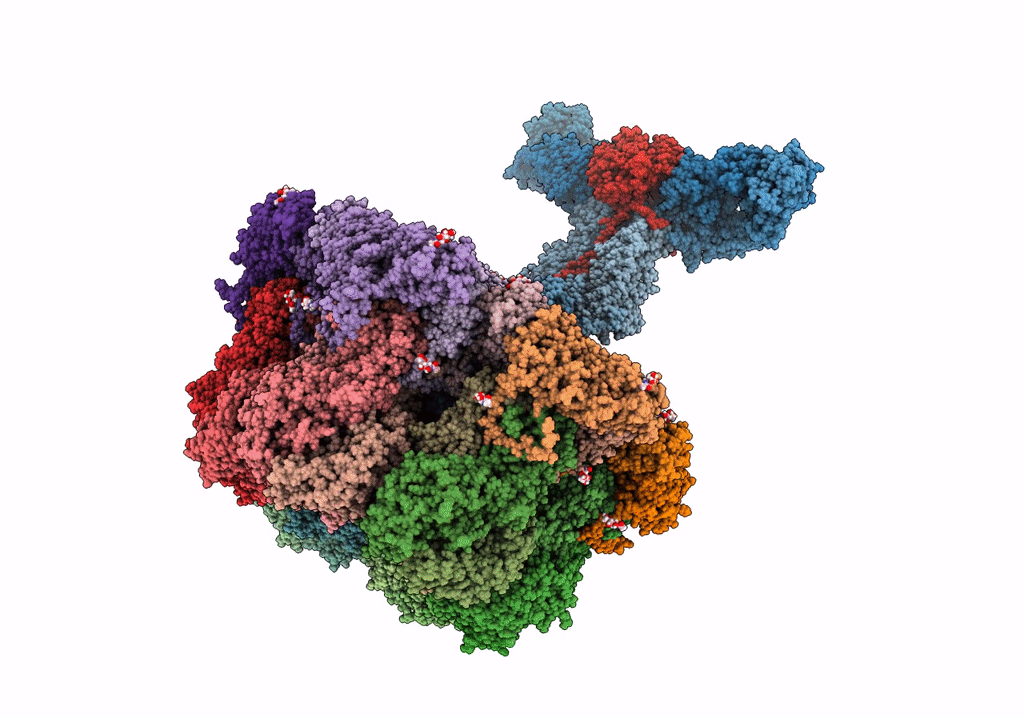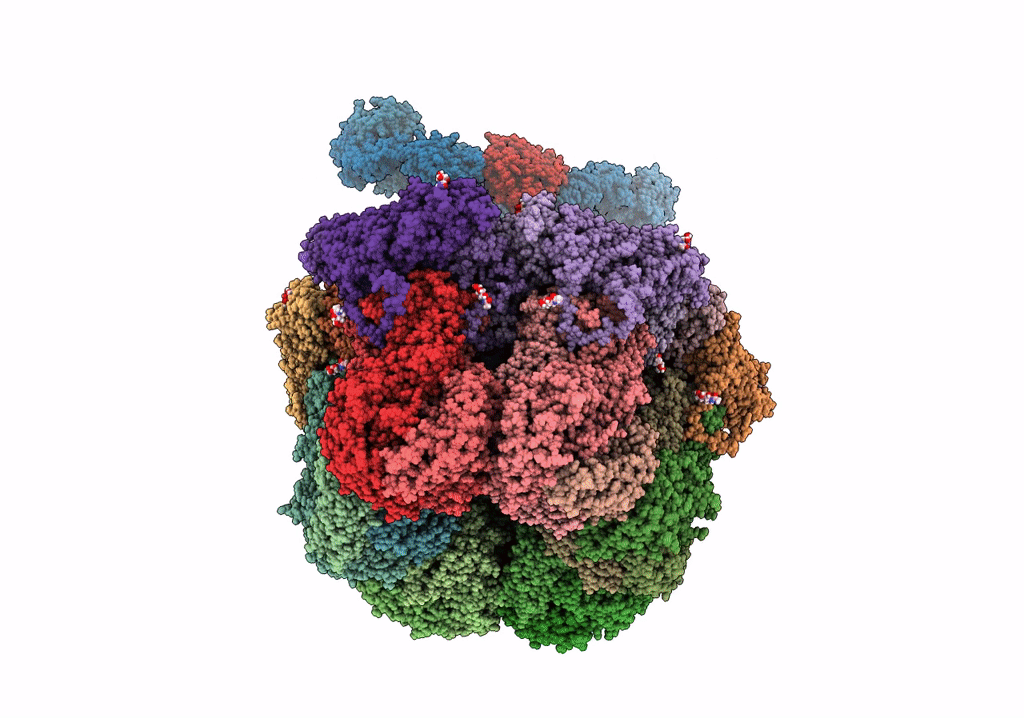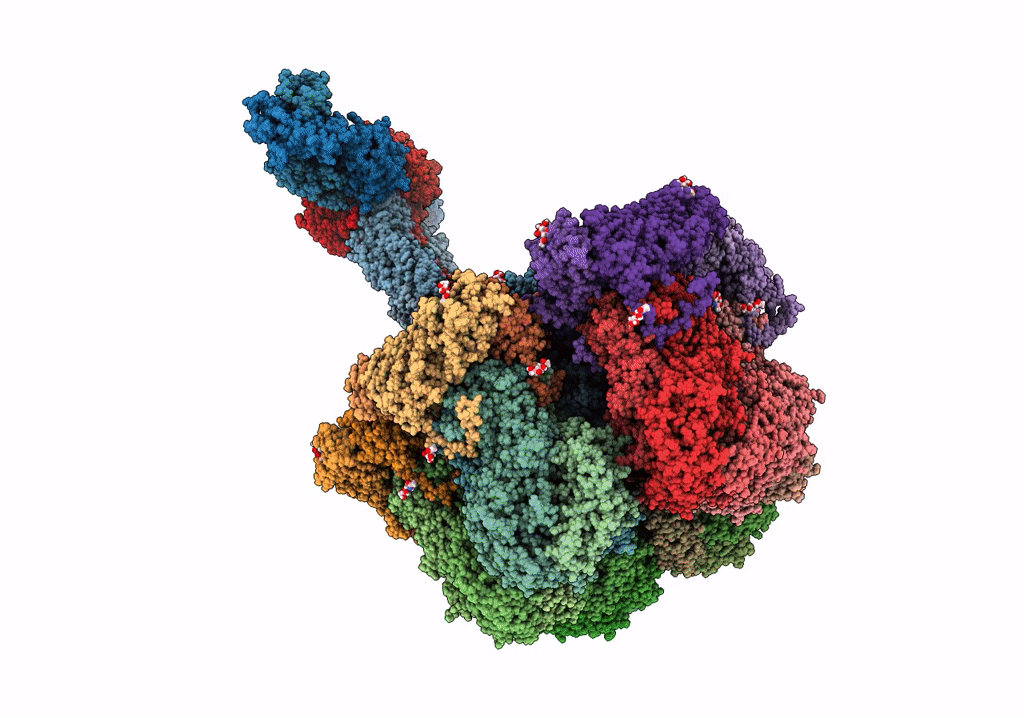 |
Structure Of The Vp5*/Vp8* Assembly From The Human Rotavirus Strain Cdc-9 In Complex With Antibody 41 - Upright Conformation
Organism: Rotavirus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-07-27 Classification: VIRUS Ligands: NAG, CA |
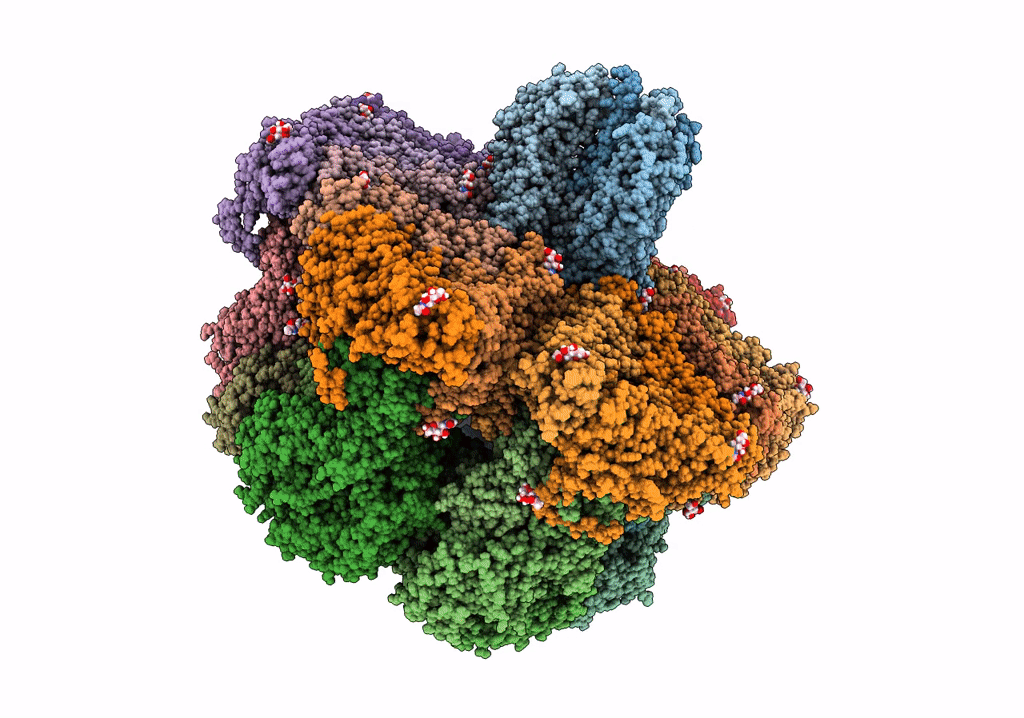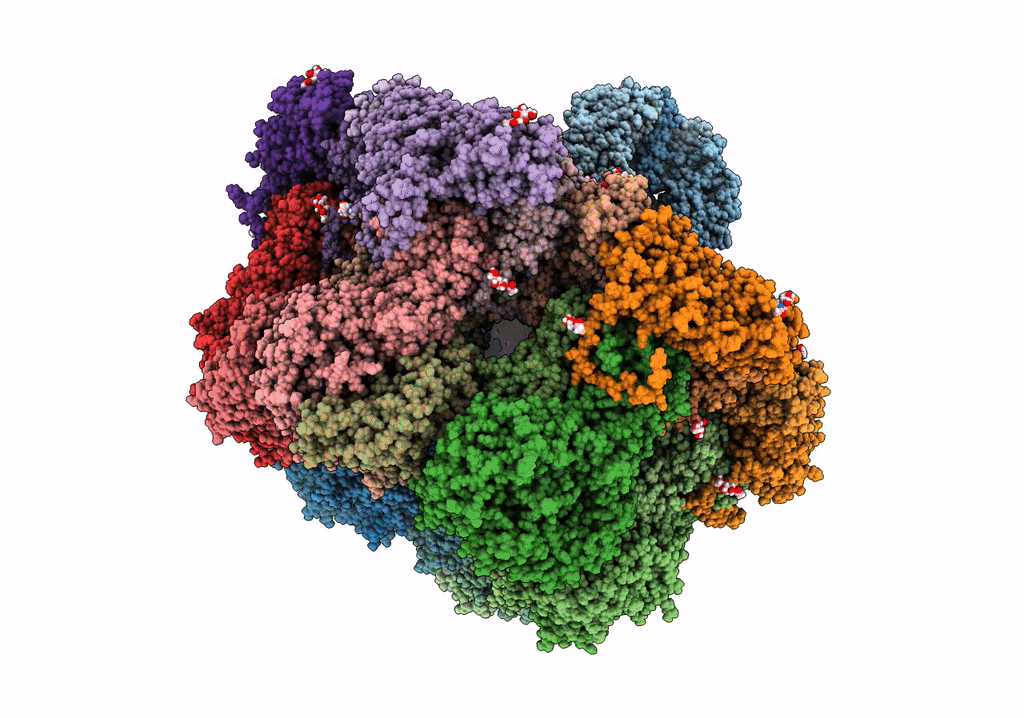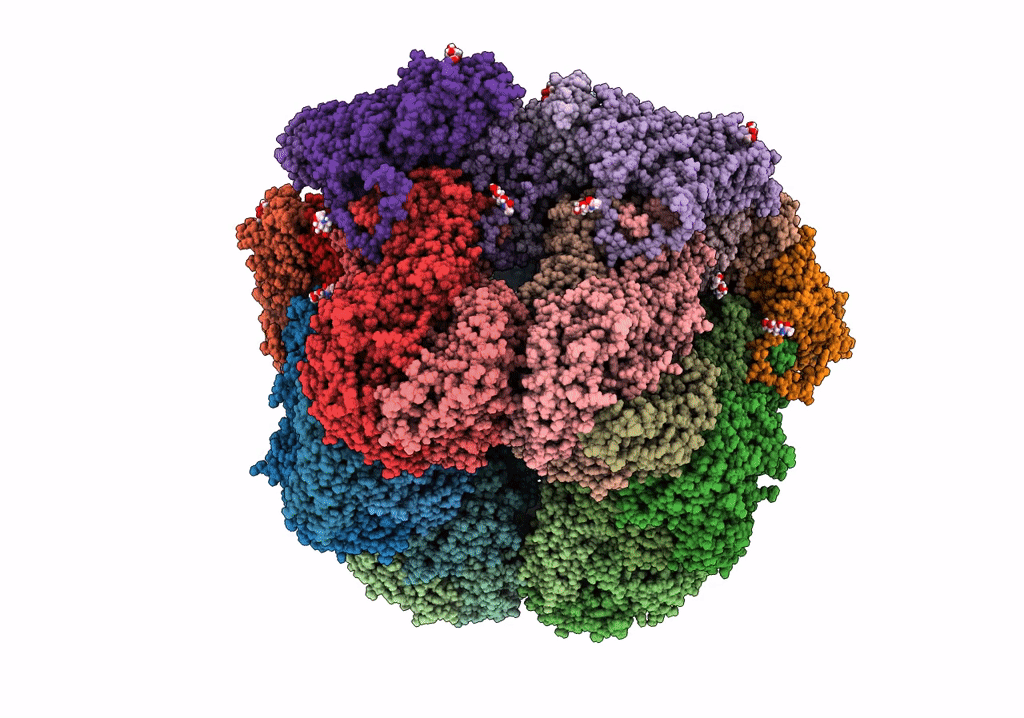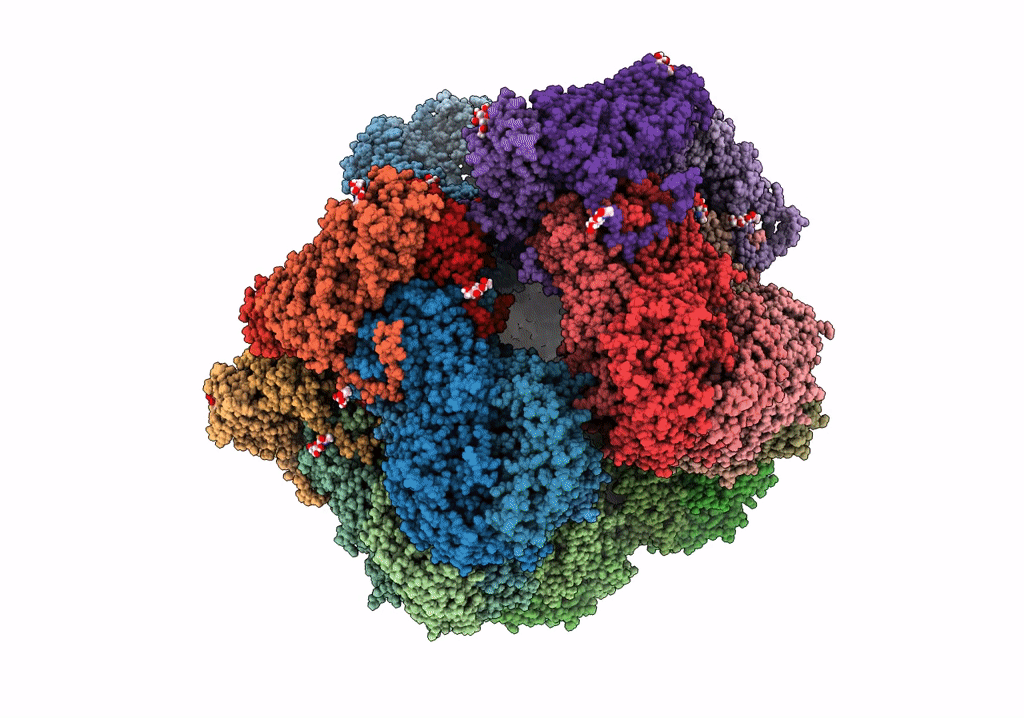 |
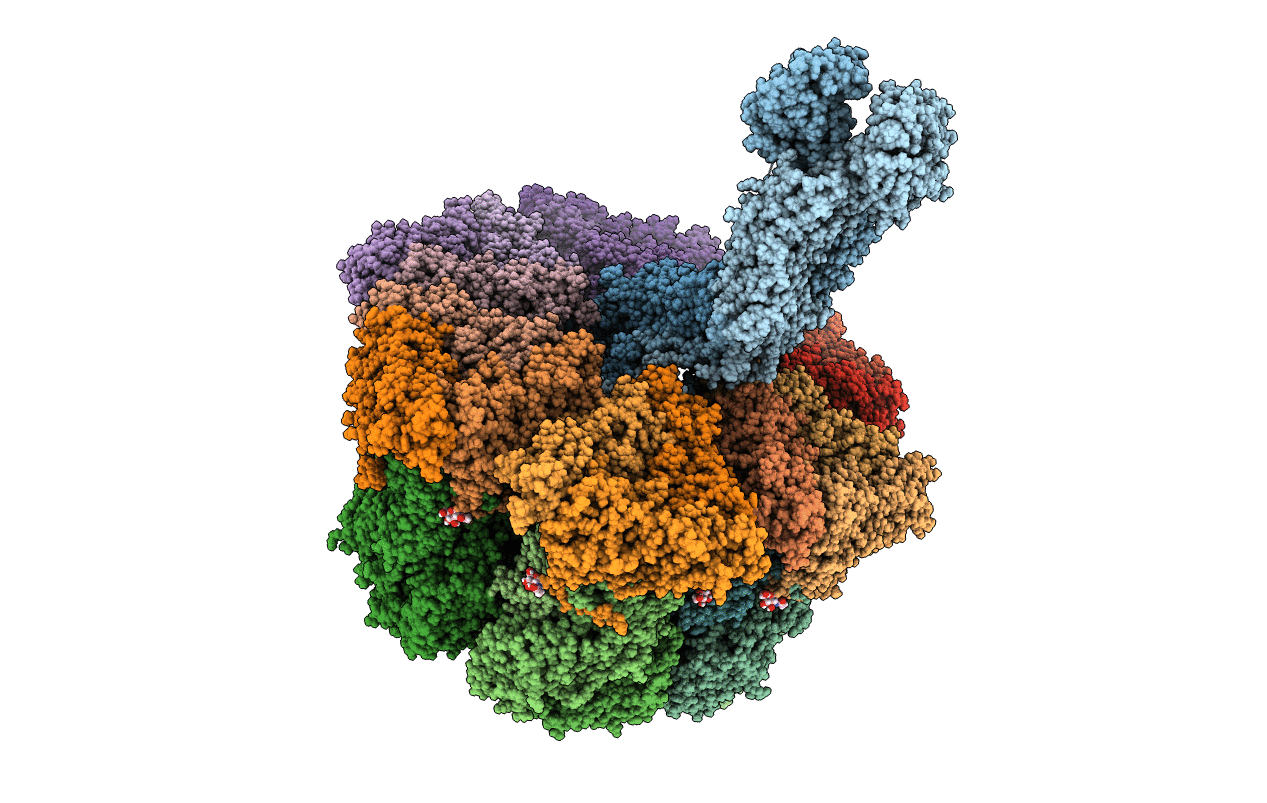
Structure Of The Vp5*/Vp8* Assembly From The Human Rotavirus Strain Cdc-9 - Reversed Conformation
Organism: Rotavirus
Method: ELECTRON MICROSCOPY Release Date: 2022-07-27 Classification: VIRUS Ligands: NAG, CA |
 |
Cryo-Em Reconstruction Of Vp5*/Vp8* Assembly From Rhesus Rotavirus Particles - Upright Conformation
Organism: Rotavirus a (strain rva/monkey/united states/rrv/1975/g3p5b[3])
Method: ELECTRON MICROSCOPY Release Date: 2021-01-20 Classification: VIRAL PROTEIN Ligands: NAG, CA |
 |
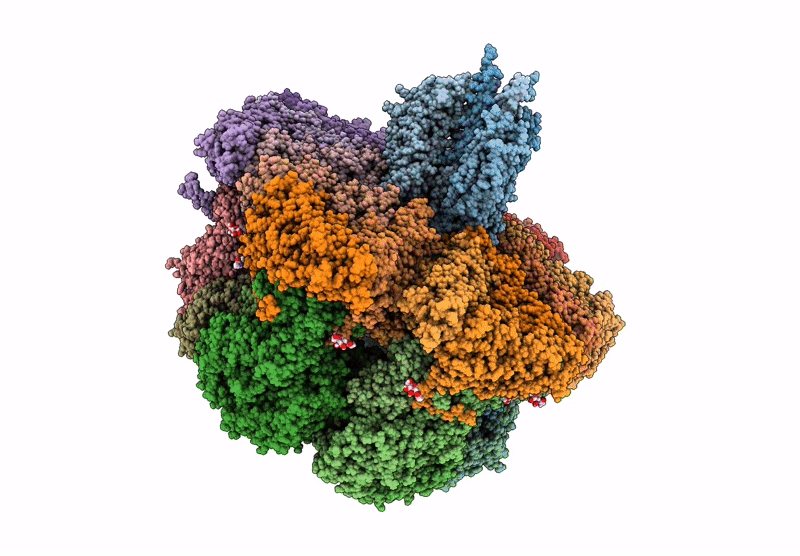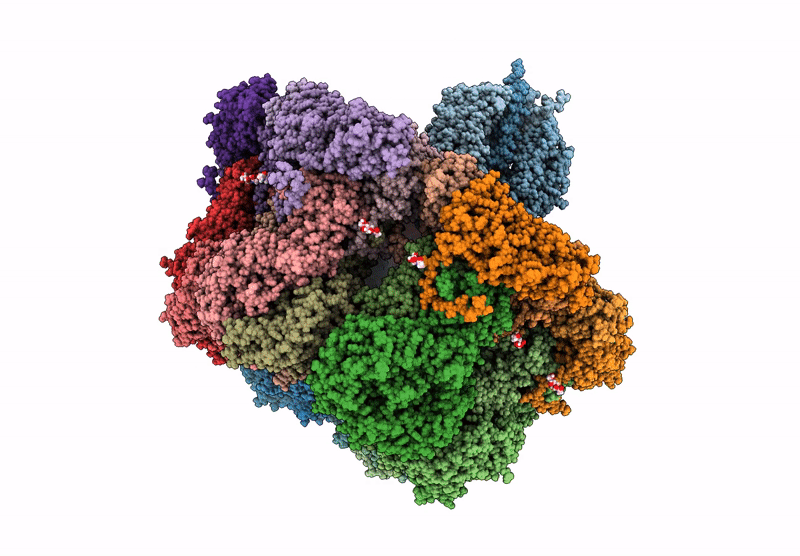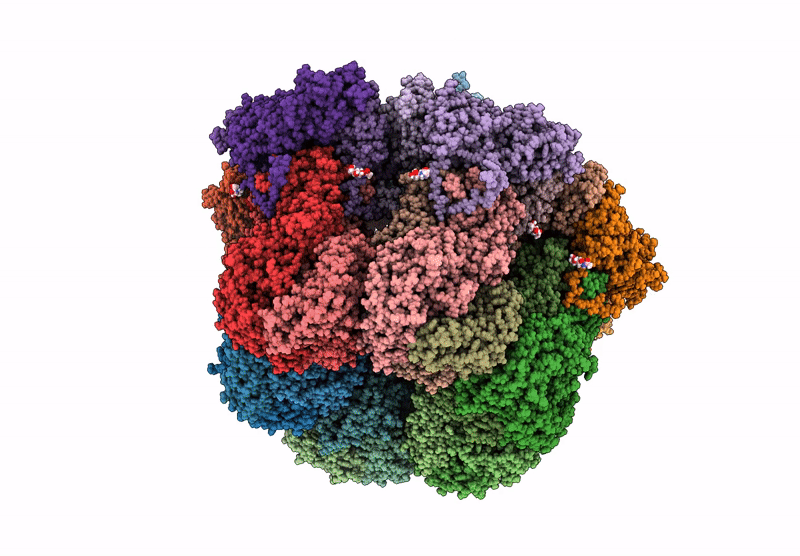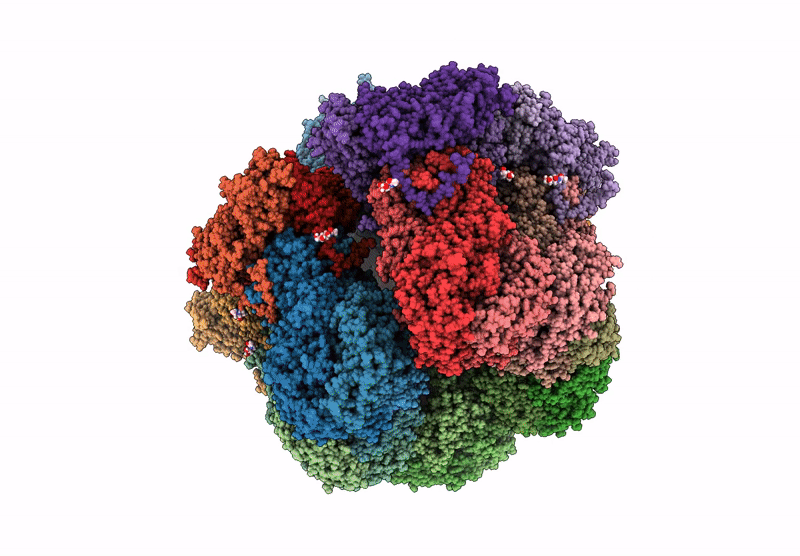
Cryo-Em Reconstruction Of Vp5*/Vp8* Assembly From Rhesus Rotavirus Particles - Intermediate Conformation
Organism: Rotavirus a (strain rva/monkey/united states/rrv/1975/g3p5b[3])
Method: ELECTRON MICROSCOPY Release Date: 2021-01-20 Classification: VIRAL PROTEIN Ligands: NAG, CA |
 |
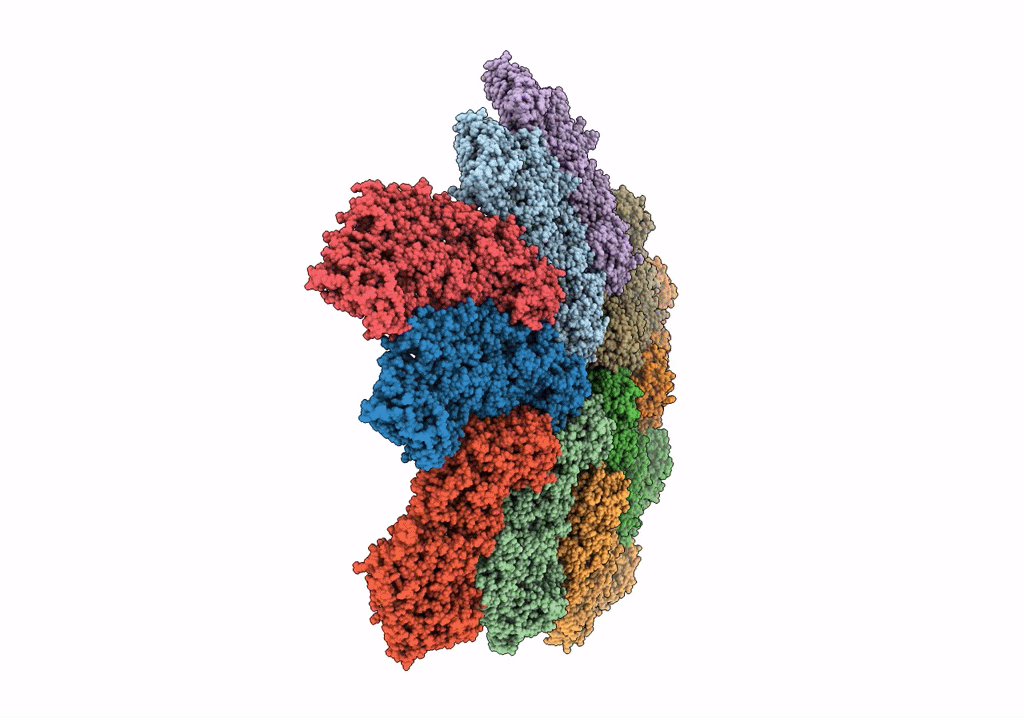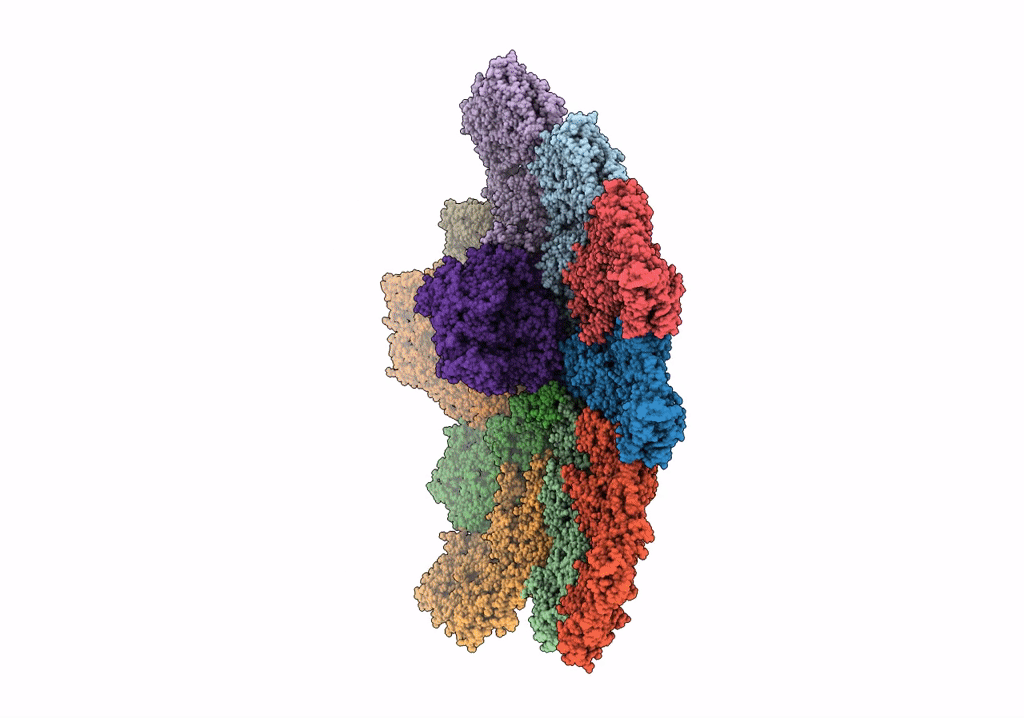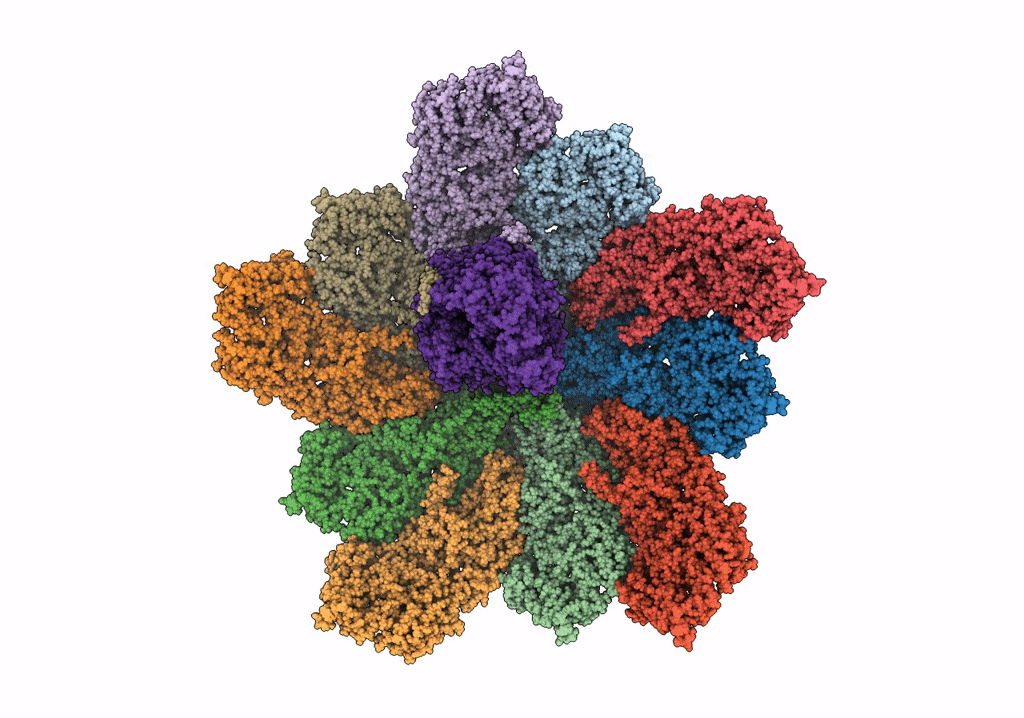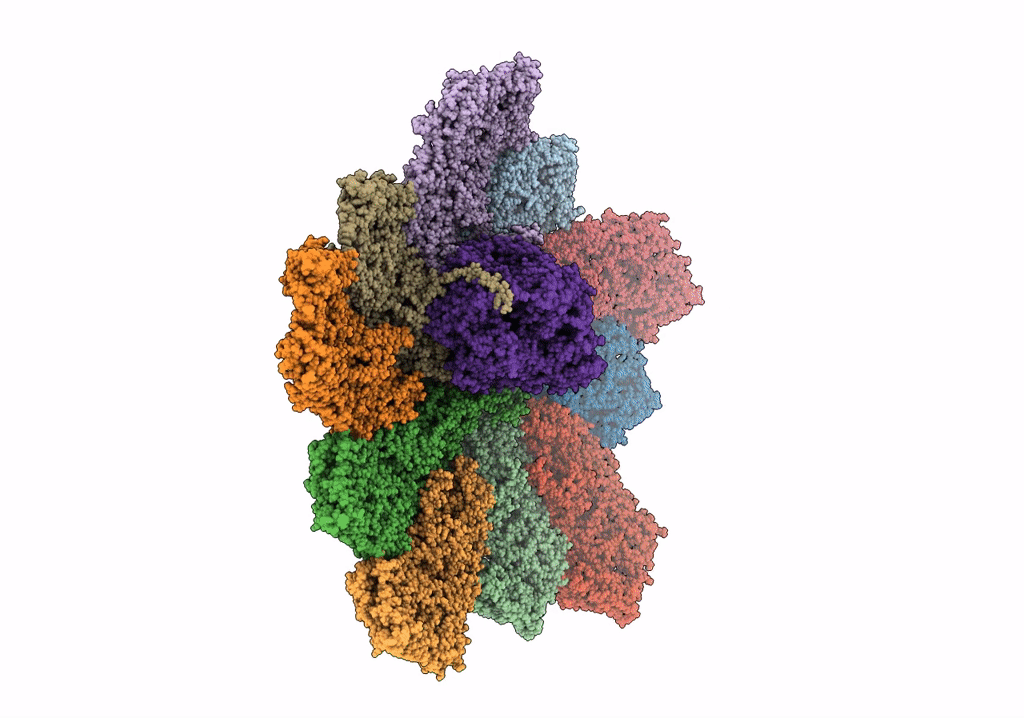
Cryo-Em Reconstruction Of Vp5*/Vp8* Assembly From Rhesus Rotavirus Particles - Reversed Conformation
Organism: Rotavirus a (strain rva/monkey/united states/rrv/1975/g3p5b[3])
Method: ELECTRON MICROSCOPY Release Date: 2021-01-20 Classification: VIRAL PROTEIN Ligands: NAG, CA |
 |
Organism: Rotavirus a (strain rva/monkey/united states/rrv/1975/g3p5b[3])
Method: ELECTRON MICROSCOPY Release Date: 2019-04-24 Classification: VIRAL PROTEIN/TRANSFERASE |
 |
Organism: Rotavirus a (strain rva/monkey/united states/rrv/1975/g3p5b[3])
Method: ELECTRON MICROSCOPY Release Date: 2019-04-24 Classification: VIRAL PROTEIN/TRANSFERASE |
 |
Organism: Rotavirus a (strain rva/monkey/united states/rrv/1975/g3p5b[3])
Method: ELECTRON MICROSCOPY Release Date: 2019-04-24 Classification: VIRAL PROTEIN/TRANSFERASE |
 |
Organism: Rotavirus a (strain rva/monkey/united states/rrv/1975/g3p5b[3])
Method: ELECTRON MICROSCOPY Release Date: 2019-04-24 Classification: VIRAL PROTEIN/TRANSFERASE/RNA |
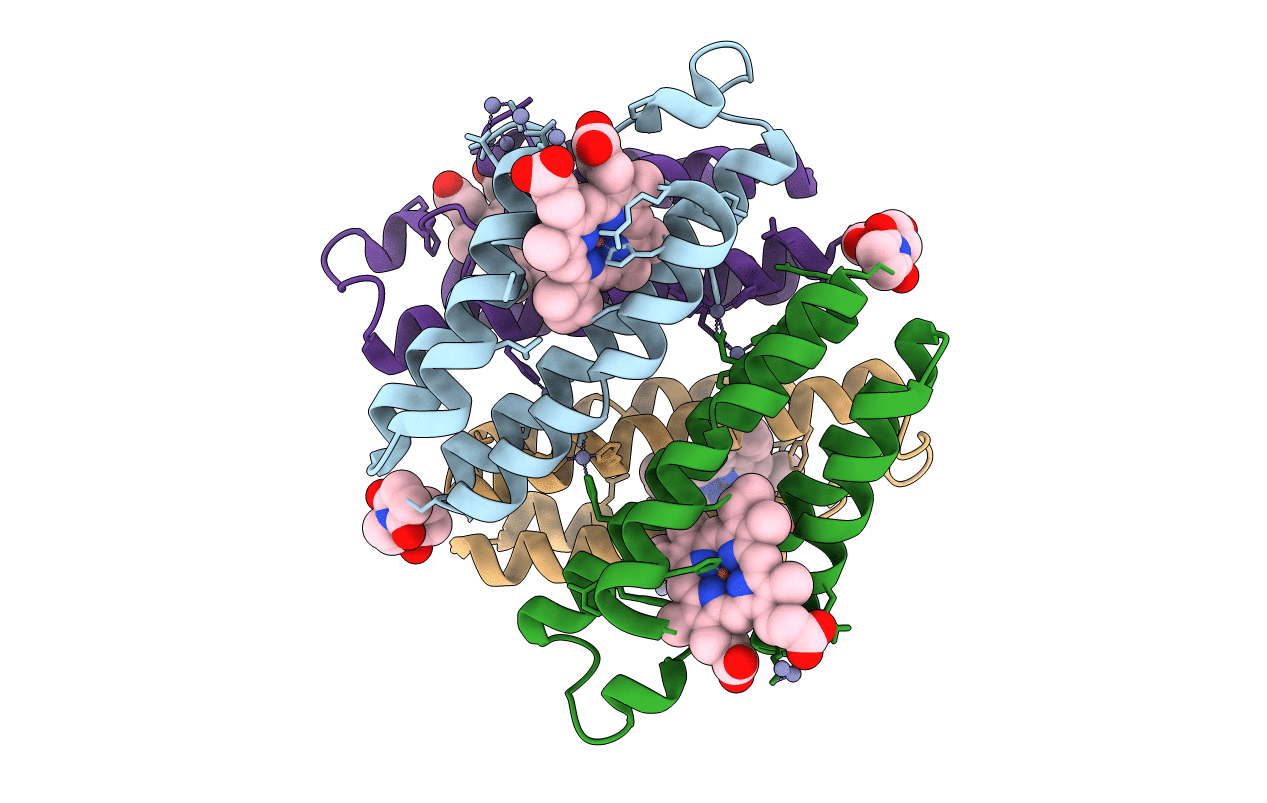 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2011-06-22 Classification: OXIDOREDUCTASE Ligands: HEM, ZN, ME7 |
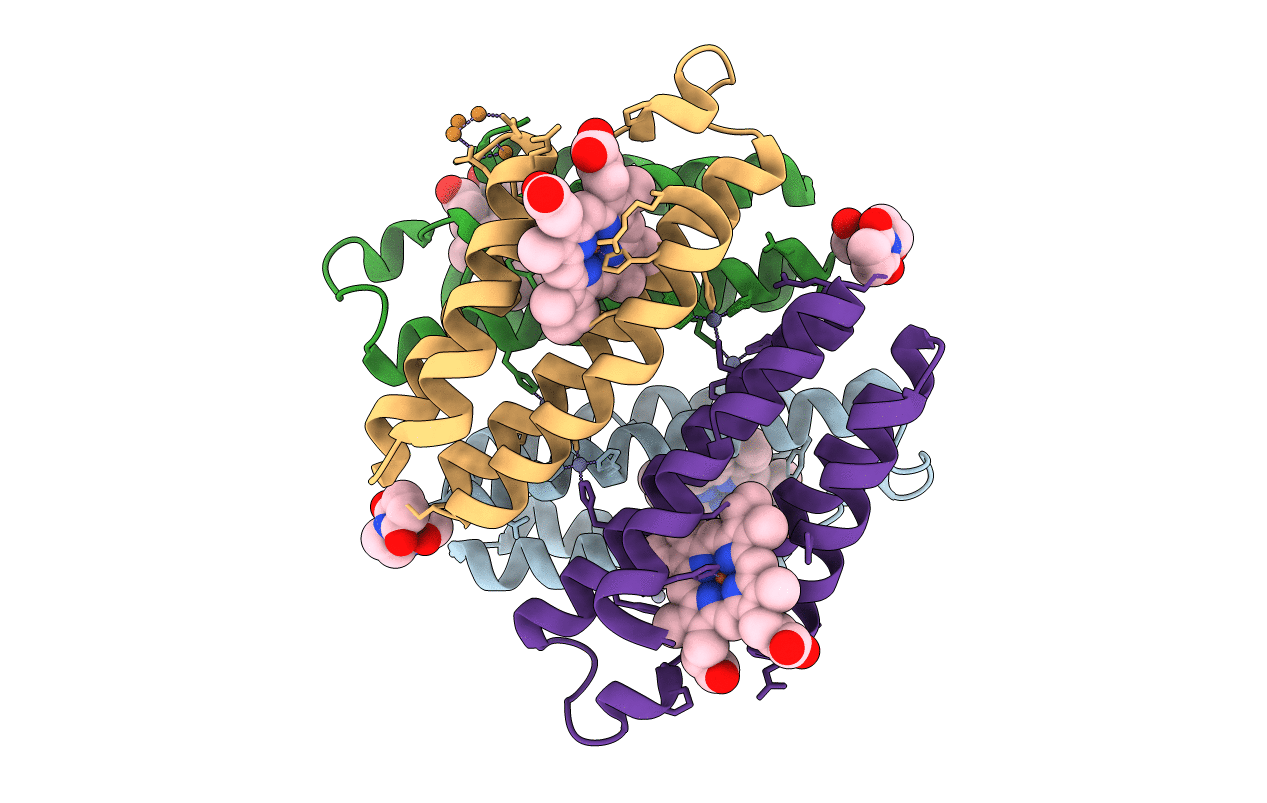 |
Crystal Structure Of The Zn-Ridc1 Complex Stabilized By Bmoe Crosslinks Cocrystallized In The Presence Of Cu(Ii)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.64 Å Release Date: 2011-06-22 Classification: OXIDOREDUCTASE Ligands: HEM, ME7, ZN, CU |
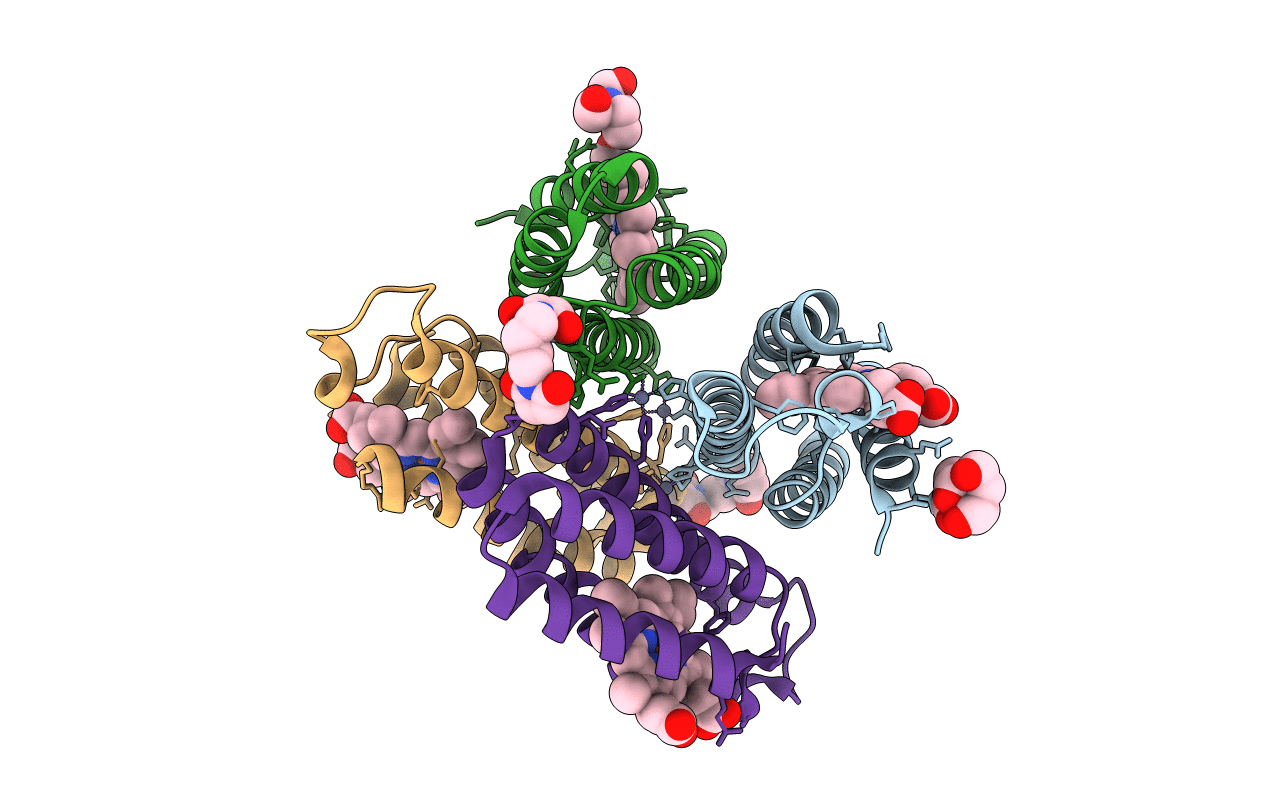 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2011-06-22 Classification: OXIDOREDUCTASE Ligands: HEM, ME9, ZN, BTB |
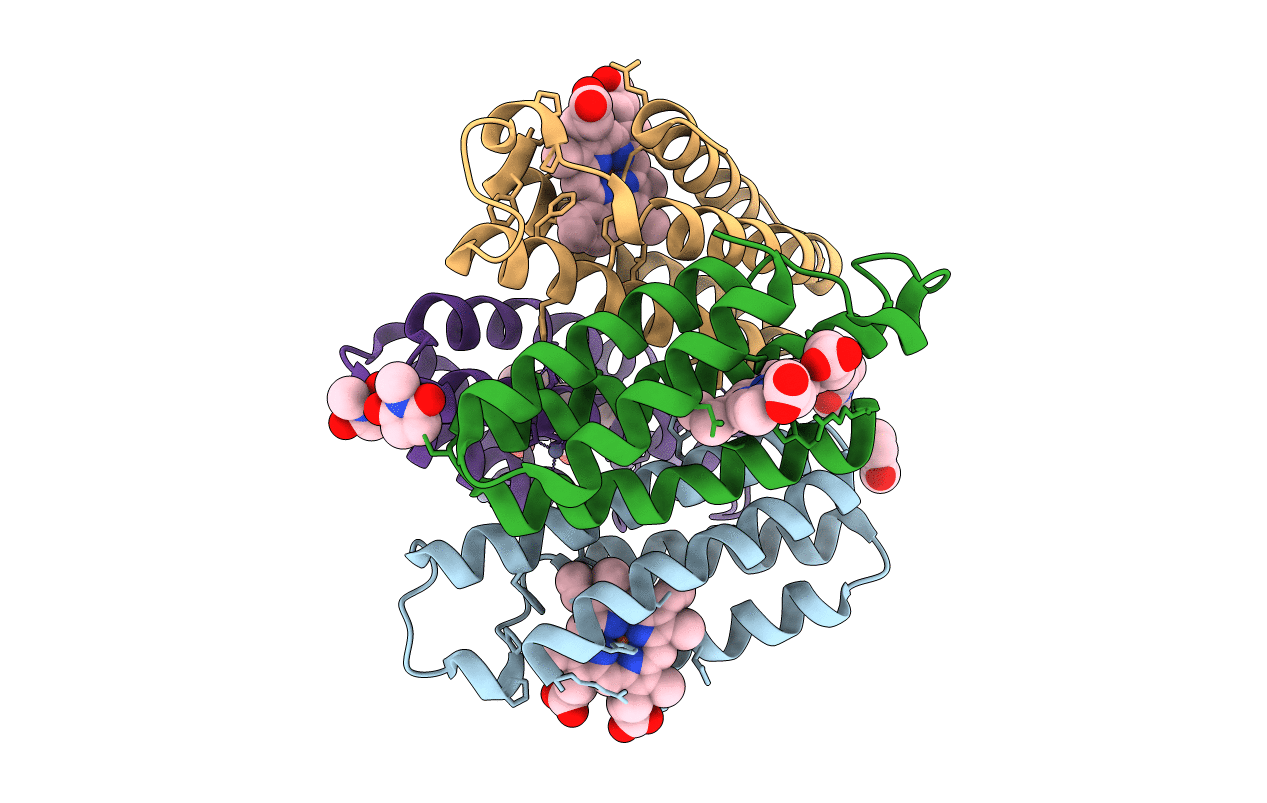 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2011-06-22 Classification: OXIDOREDUCTASE Ligands: ZN, HEM, LCY |
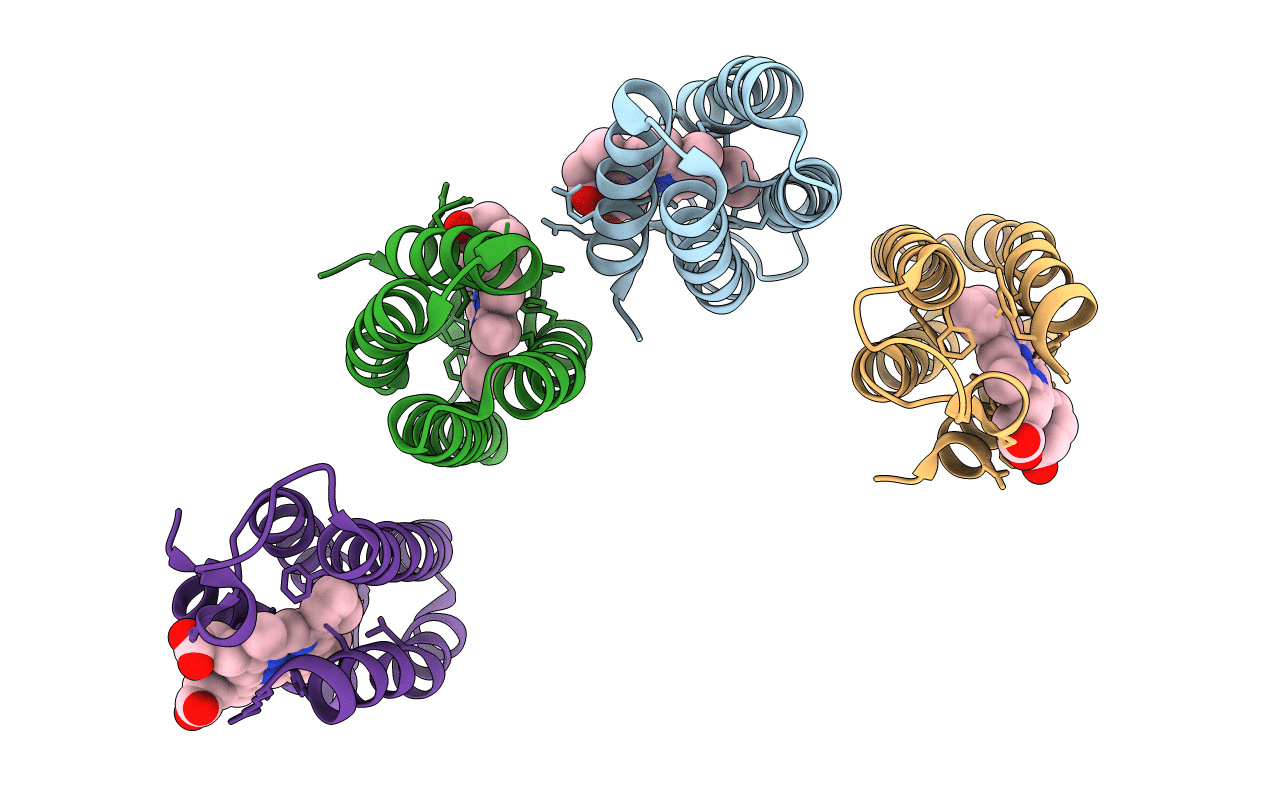 |
Crystal Structure Of An Engineered Metal-Free Tetrameric Cytochrome Cb562 Complex Templated By Zn-Coordination
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2010-06-16 Classification: ELECTRON TRANSPORT Ligands: HEM |
 |
Crystal Structure Of A Tetrameric Zn-Bound Cytochrome Cb562 Complex With Covalently And Non-Covalently Stabilized Interfaces
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2010-06-16 Classification: ELECTRON TRANSPORT Ligands: HEM, ZN |
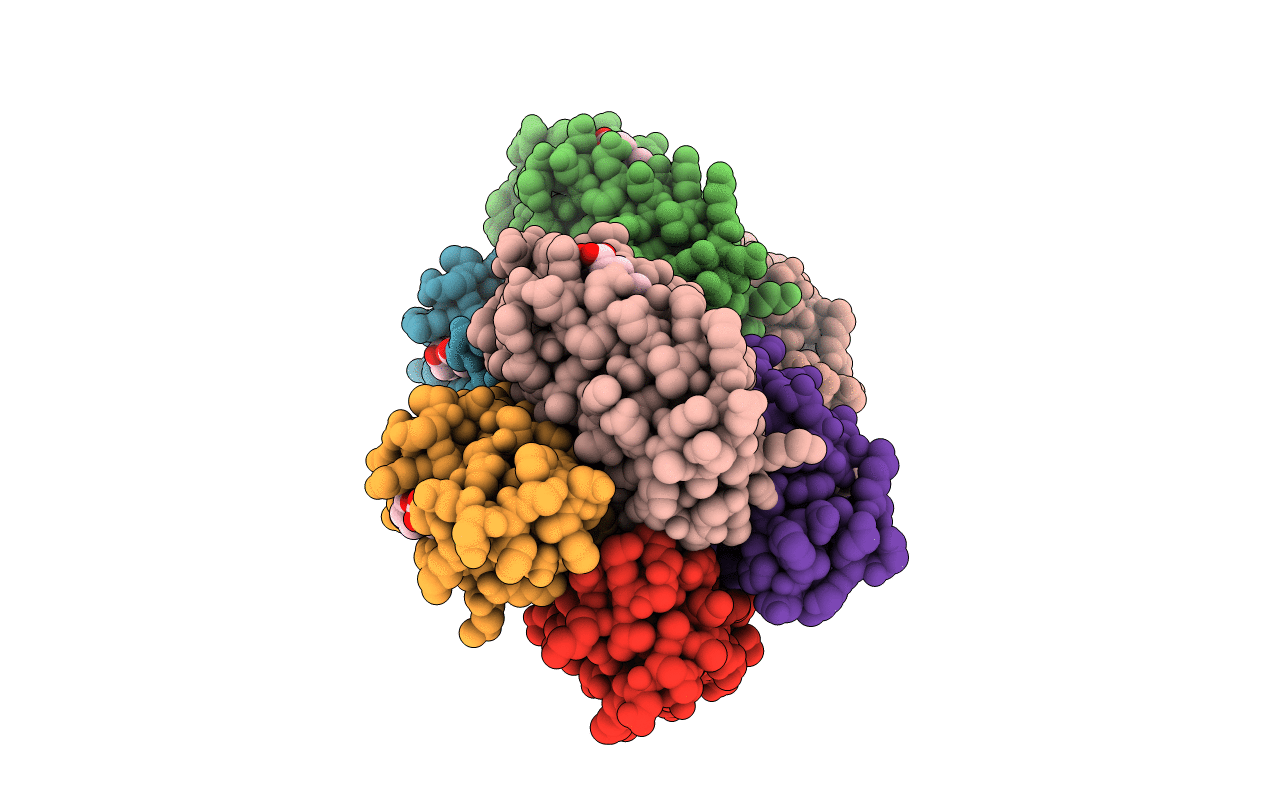 |
A Tetrameric Zn-Bound Cytochrome Cb562 Complex With Covalently And Non-Covalently Stabilized Interfaces Crystallized In The Presence Of Cu(Ii) And Zn(Ii)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2010-06-16 Classification: OXIDOREDUCTASE Ligands: HEM, ZN |
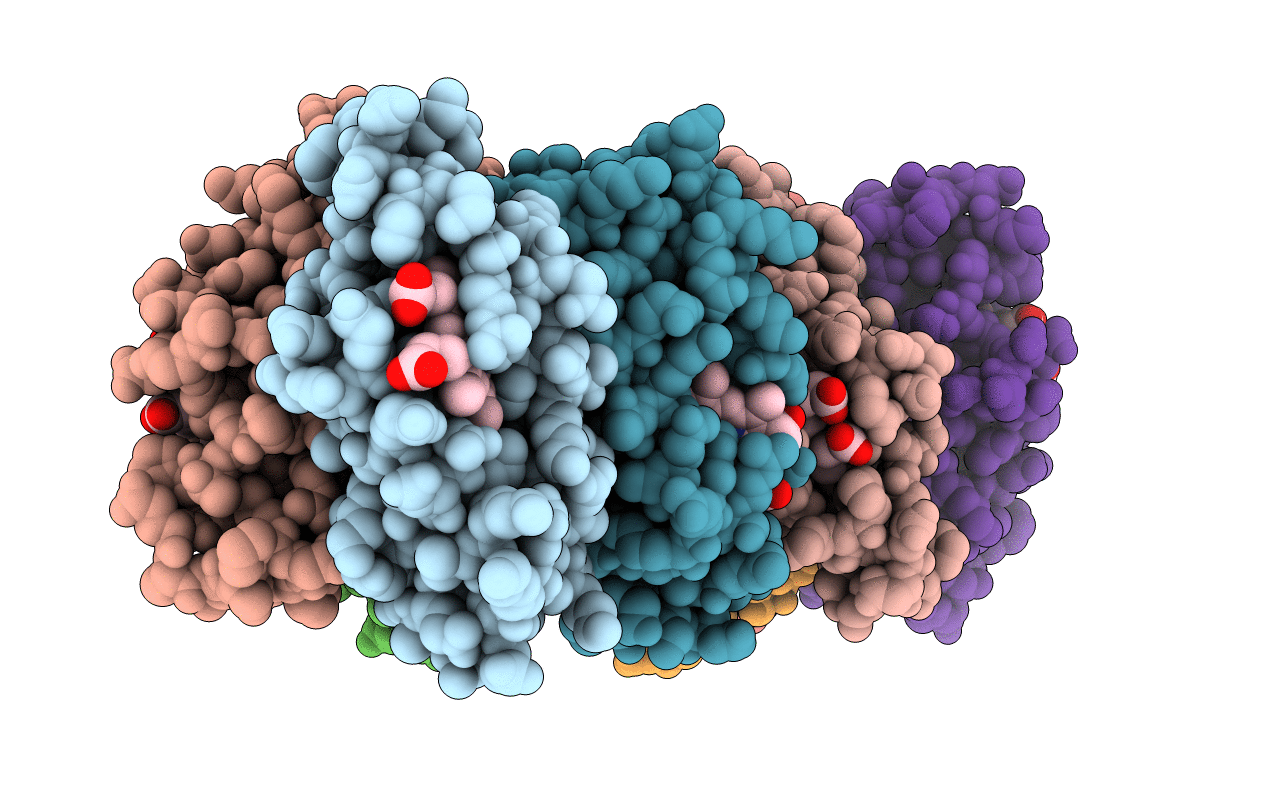 |
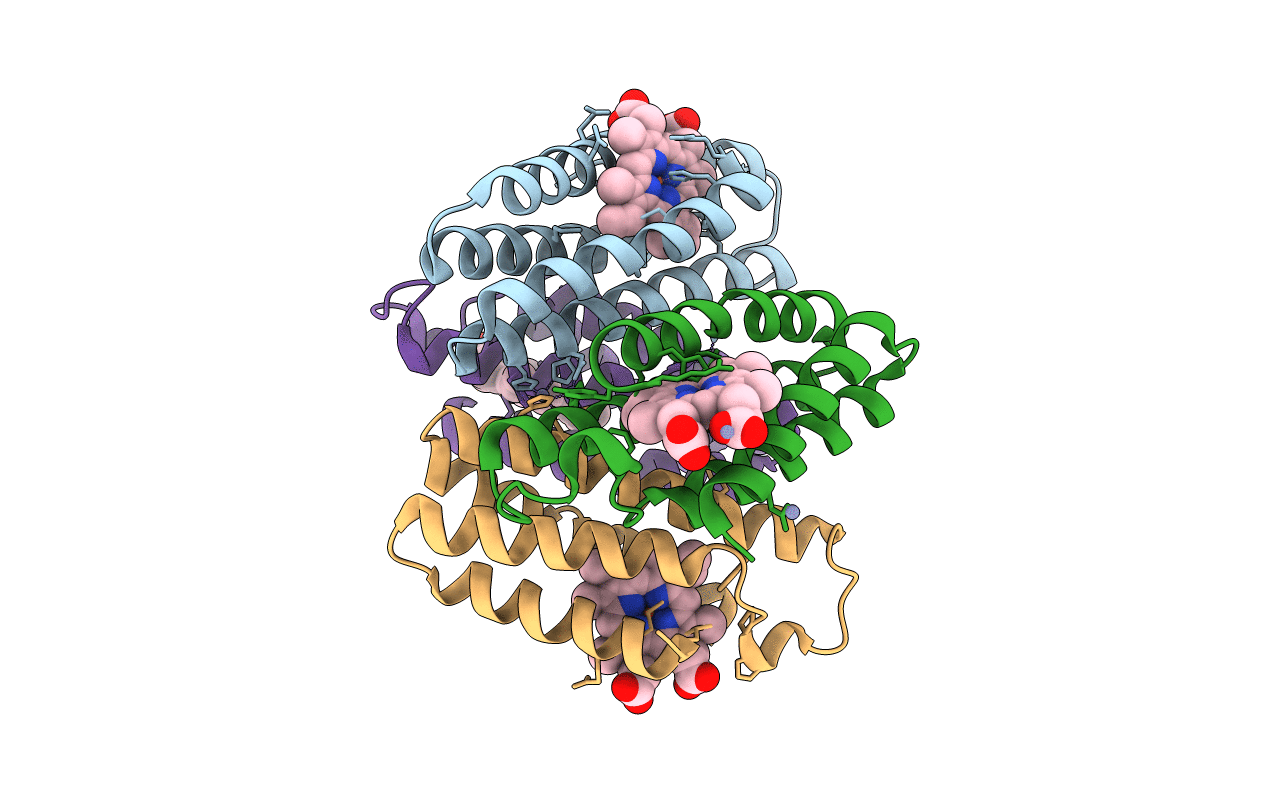
Crystal Structure Of The Zn-Induced Tetramer Of The Engineered Cyt Cb562 Variant Ridc-1
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2010-02-09 Classification: METAL BINDING PROTEIN Ligands: HEM, ZN |
 |
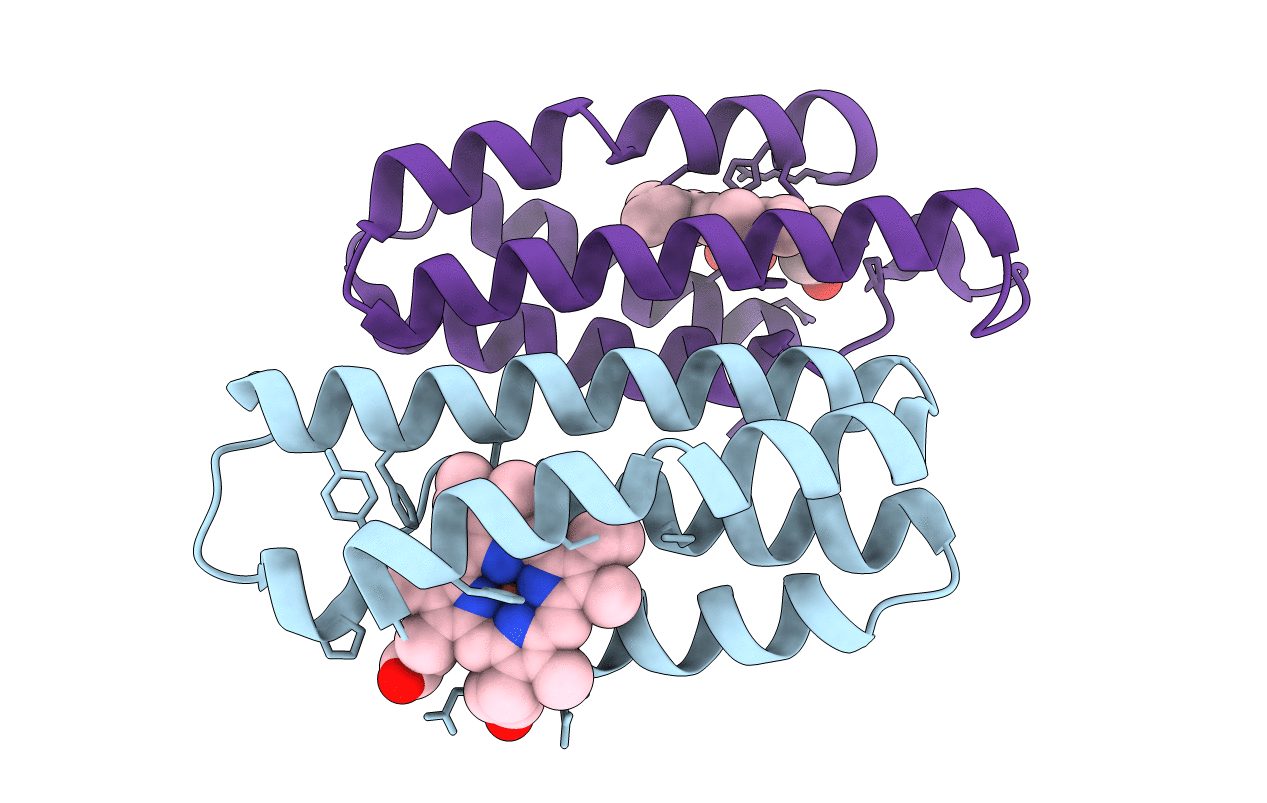
Crystal Structure Of The Zn-Induced Tetramer Of The Engineered Cyt Cb562 Variant Ridc-2
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2010-02-09 Classification: METAL BINDING PROTEIN Ligands: HEM, ZN |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2010-02-09 Classification: METAL BINDING PROTEIN Ligands: HEM |
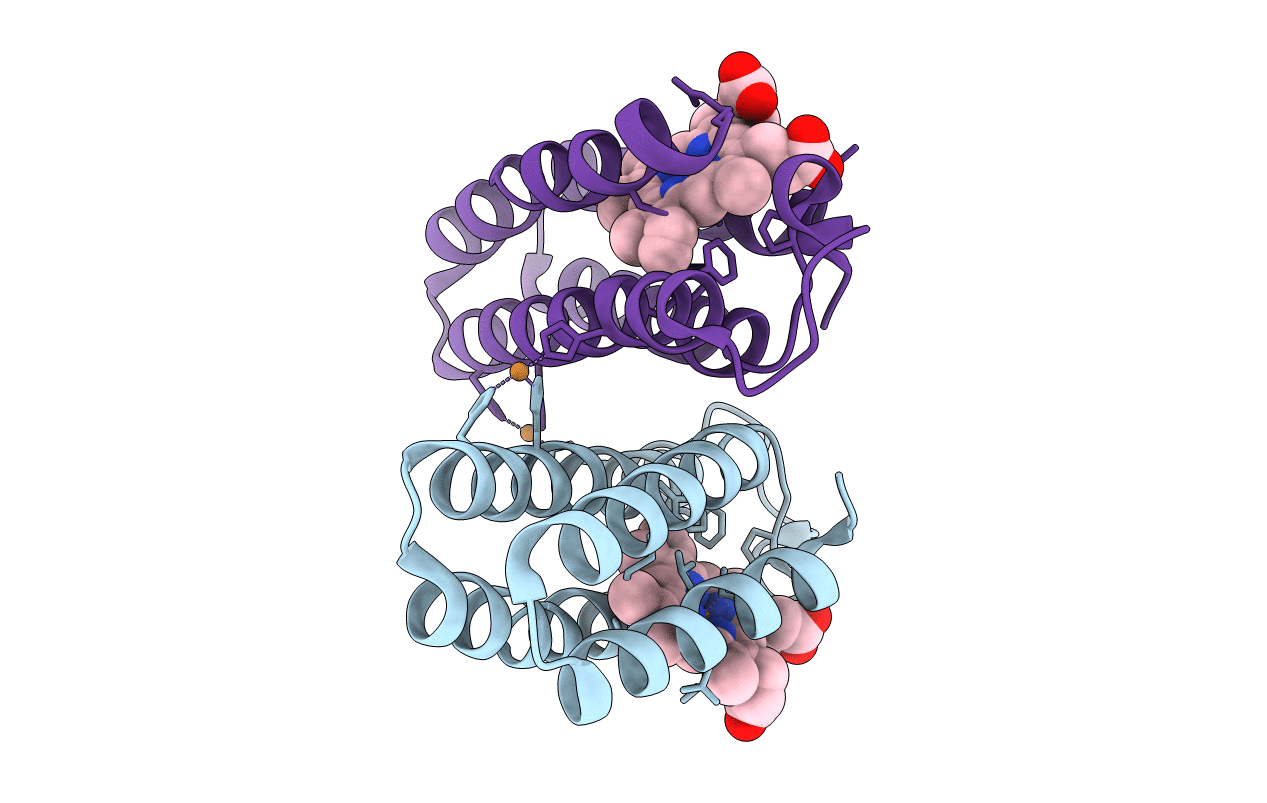 |
Crystal Structure Of The Cu-Induced Dimer Of The Engineered Cyt Cb562 Variant Ridc-1
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2010-02-09 Classification: METAL BINDING PROTEIN Ligands: HEM, CU, CL |

