Search Count: 17
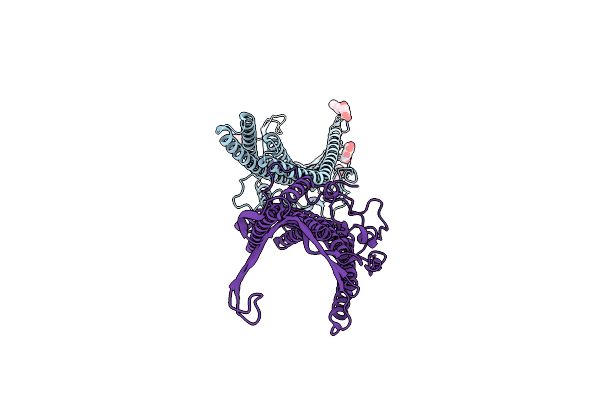 |
Deciphering The Role Of The Channel Constrictions In The Opening Mechanism Of Mexab-Oprm Efflux Pump From Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2022-04-13 Classification: TRANSLOCASE Ligands: PLM, BOG |
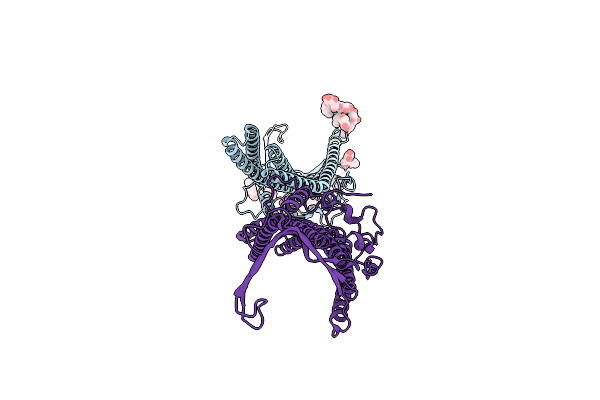 |
Deciphering The Role Of The Channel Constrictions In The Opening Mechanism Of Mexab-Oprm Efflux Pump From Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2021-07-28 Classification: TRANSLOCASE Ligands: PLM, SO4, BOG |
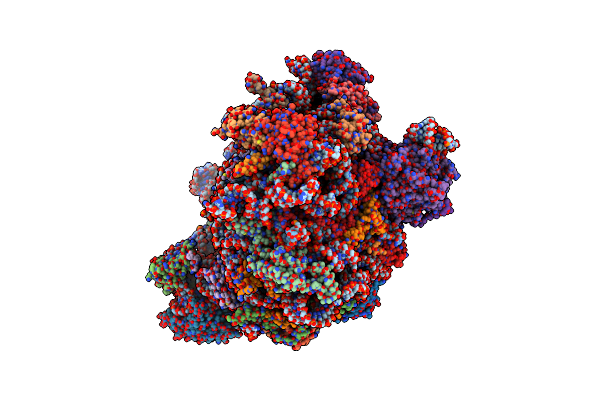 |
Structure Of The Yeast 60S Ribosomal Subunit In Complex With Arx1, Alb1 And N-Terminally Tagged Rei1
Organism: Saccharomyces cerevisiae s288c, Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2015-12-16 Classification: RIBOSOME Ligands: MG, ZN |
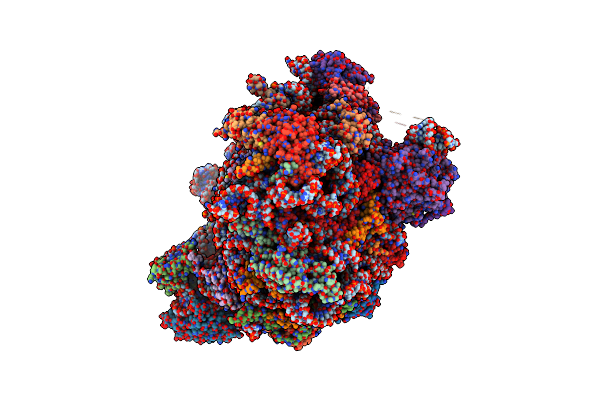 |
Structure Of The Yeast 60S Ribosomal Subunit In Complex With Arx1, Alb1 And C-Terminally Tagged Rei1
Organism: Saccharomyces cerevisiae s288c, Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Resolution:3.41 Å Release Date: 2015-12-16 Classification: RIBOSOME Ligands: MG, ZN |
 |
Organism: Bacillus amyloliquefaciens
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2010-03-09 Classification: HYDROLASE Ligands: PTD |
 |
Organism: Escherichia coli o6
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2007-05-22 Classification: TRANSFERASE Ligands: ADP |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2007-05-22 Classification: TRANSFERASE Ligands: AFH |
 |
Organism: Escherichia coli uti89
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2007-05-01 Classification: TRANSFERASE |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2006-04-25 Classification: HYDROLASE Ligands: NO3 |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2006-04-25 Classification: HYDROLASE Ligands: NO3, URE |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2006-04-25 Classification: HYDROLASE Ligands: NO3, ACT, TOU |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2006-04-25 Classification: HYDROLASE Ligands: NO3, BRJ |
 |
Organism: Bacillus amyloliquefaciens
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2006-04-25 Classification: HYDROLASE Ligands: ZN |
 |
Organism: Bacillus amyloliquefaciens
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2006-04-25 Classification: HYDROLASE Ligands: URE, ZN |
 |
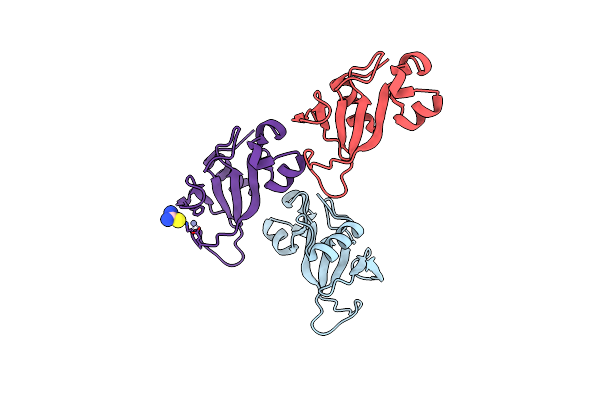
Organism: Bacillus amyloliquefaciens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2006-04-25 Classification: HYDROLASE Ligands: BRJ |
 |
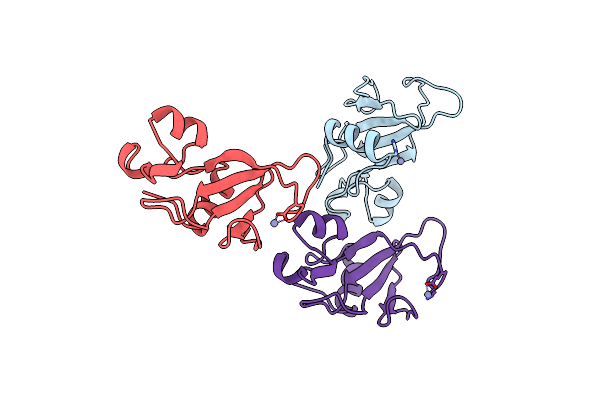
Organism: Bacillus amyloliquefaciens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2006-04-25 Classification: HYDROLASE Ligands: ZN, TOU |
 |
Organism: Bacillus amyloliquefaciens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 1998-04-29 Classification: RIBONUCLEASE Ligands: ZN |

