Search Count: 36
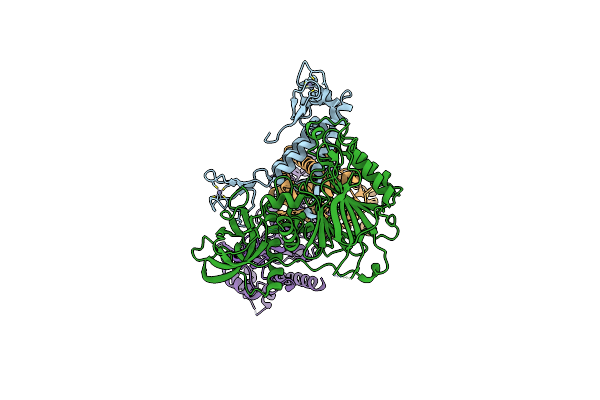 |
Tetramer Core Subcomplex (Conformation 1) Of Xenopus Laevis Dna Polymerase Alpha-Primase
Organism: Xenopus laevis
Method: ELECTRON MICROSCOPY Release Date: 2023-12-20 Classification: REPLICATION, TRANSFERASE Ligands: ZN |
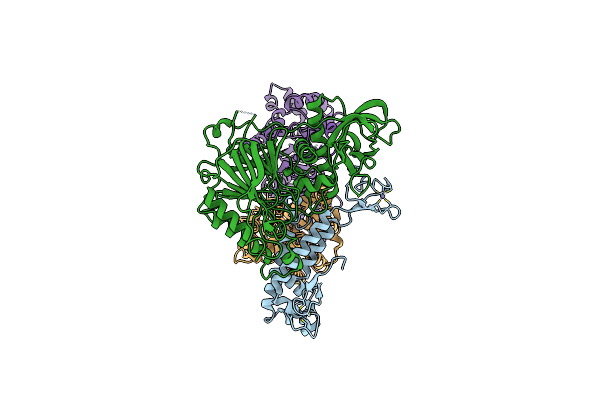 |
Tetramer Core Subcomplex (Conformation 2) Of Xenopus Laevis Dna Polymerase Alpha-Primase
Organism: Xenopus laevis
Method: ELECTRON MICROSCOPY Release Date: 2023-12-20 Classification: REPLICATION, TRANSFERASE Ligands: ZN |
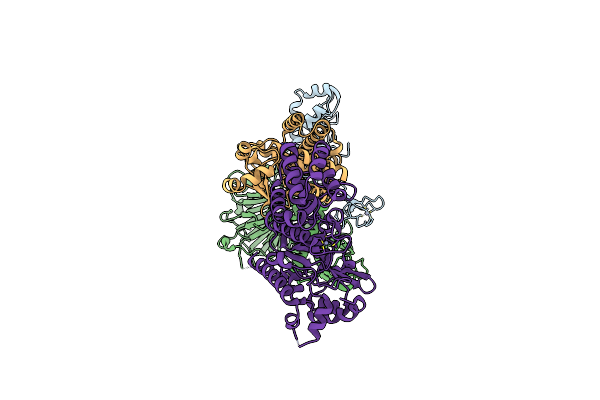 |
Tetramer Core Subcomplex (Conformation 3) Of Xenopus Laevis Dna Polymerase Alpha-Primase
Organism: Xenopus laevis
Method: ELECTRON MICROSCOPY Release Date: 2023-12-20 Classification: REPLICATION, TRANSFERASE Ligands: ZN |
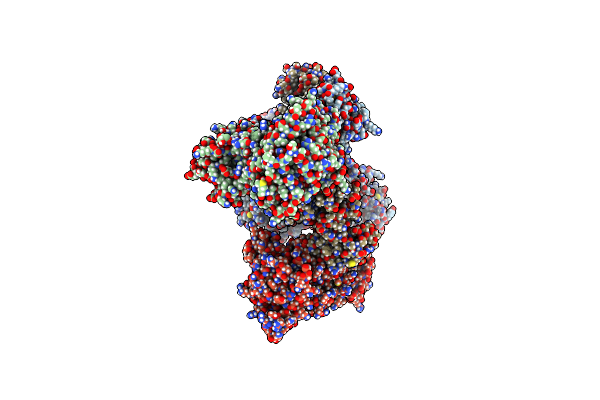 |
Dna Initiation Complex (Configuration 1) Of Xenopus Laevis Dna Polymerase Alpha-Primase
Organism: Xenopus laevis, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-12-20 Classification: REPLICATION, TRANSFERASE Ligands: ZN, DGT, MG, SF4 |
 |
Dna Initiation Complex (Configuration 2) Of Xenopus Laevis Dna Polymerase Alpha-Primase
Organism: Xenopus laevis, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-12-20 Classification: REPLICATION, TRANSFERASE/DNA/RNA Ligands: ZN, DGT, MG, SF4 |
 |
Dna Elongation Complex (Configuration 1) Of Xenopus Laevis Dna Polymerase Alpha-Primase
Organism: Xenopus laevis, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-12-20 Classification: REPLICATION, TRANSFERASE/DNA/RNA Ligands: ZN, DGT, MG, SF4 |
 |
Dna Elongation Complex (Configuration 2) Of Xenopus Laevis Dna Polymerase Alpha-Primase
Organism: Xenopus laevis, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-12-20 Classification: REPLICATION, TRANSFERASE/DNA/RNA Ligands: ZN, DGT, MG, SF4 |
 |
Partial Dna Termination Subcomplex Of Xenopus Laevis Dna Polymerase Alpha-Primase
Organism: Xenopus laevis, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-10-11 Classification: REPLICATION, TRANSFERASE/DNA/RNA Ligands: DGT, MG |
 |
Complete Dna Termination Subcomplex 1 Of Xenopus Laevis Dna Polymerase Alpha-Primase
Organism: Xenopus laevis, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-10-11 Classification: REPLICATION, TRANSFERASE/DNA/RNA Ligands: DGT, MG, SF4 |
 |
Complete Dna Termination Subcomplex 2 Of Xenopus Laevis Dna Polymerase Alpha-Primase
Organism: Xenopus laevis, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-10-11 Classification: REPLICATION, TRANSFERASE/DNA/RNA Ligands: DGT, MG |
 |
Partial Auto-Inhibitory Complex Of Xenopus Laevis Dna Polymerase Alpha-Primase
Organism: Xenopus laevis
Method: ELECTRON MICROSCOPY Release Date: 2023-04-12 Classification: REPLICATION, TRANSFERASE Ligands: ZN, SF4 |
 |
Complete Auto-Inhibitory Complex Of Xenopus Laevis Dna Polymerase Alpha-Primase
Organism: Xenopus laevis
Method: ELECTRON MICROSCOPY Release Date: 2023-04-12 Classification: REPLICATION, TRANSFERASE Ligands: ZN, SF4 |
 |
Organism: Xenopus laevis, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-04-12 Classification: REPLICATION, TRANSFERASE/DNA/RNA Ligands: DGT, MG, SF4 |
 |
Partial Dna Elongation Subcomplex Of Xenopus Laevis Dna Polymerase Alpha-Primase
Organism: Xenopus laevis, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-04-12 Classification: REPLICATION, TRANSFERASE/DNA/RNA Ligands: DGT, MG |
 |
Complete Dna Elongation Subcomplex Of Xenopus Laevis Dna Polymerase Alpha-Primase
Organism: Xenopus laevis, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-04-12 Classification: REPLICATION, TRANSFERASE/DNA/RNA Ligands: DGT, MG, SF4 |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2022-06-29 Classification: REPLICATION Ligands: GOL, SF4, MPD |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2022-06-29 Classification: REPLICATION Ligands: SF4, MPD |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2022-06-29 Classification: REPLICATION Ligands: SF4, MPD |
 |
Receptor For Advanced Glycation End Products Vc1 Domain In Complex With 3-(3-(((3-(4-Carboxyphenoxy)Benzyl)Oxy)Methyl)Phenyl)-1H-Indole-2-Carboxylic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2021-12-15 Classification: SIGNALING PROTEIN Ligands: ACT, CL, Y6S |
 |
Receptor For Advanced Glycation End Products Vc1 Domain In Complex With 3-(3-((4-(4-Carboxyphenoxy)Benzyl)Oxy)Phenyl)-1H-Indole-2-Carboxylic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-07-28 Classification: SIGNALING PROTEIN Ligands: Y6P, ACT, CL |

