Search Count: 12
 |
Organism: Aplysia kurodai
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2023-11-15 Classification: HYDROLASE Ligands: NAG |
 |
Organism: Aplysia kurodai
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2023-11-15 Classification: UNKNOWN FUNCTION Ligands: GOL |
 |
Organism: Aplysia kurodai
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2023-11-15 Classification: UNKNOWN FUNCTION Ligands: GOL, ACE |
 |
Eisenia Hydrolysis-Enhancing Protein From Aplysia Kurodai Complex With Tannic Acid
Organism: Aplysia kurodai
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-11-15 Classification: UNKNOWN FUNCTION Ligands: GGP |
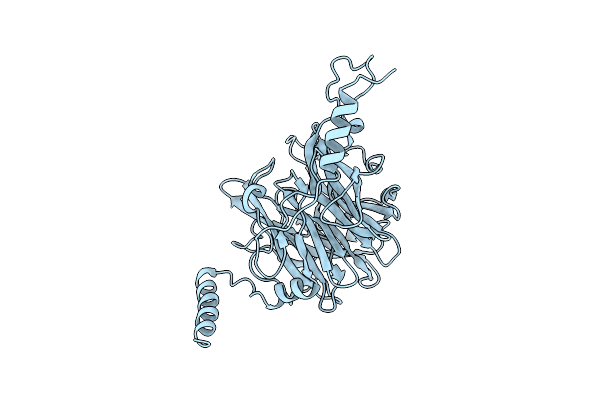 |
Crystal Structure Of Ruminococcus Albus 4-O-Beta-D-Mannosyl-D-Glucose Phosphorylase (Ramp1)
Organism: Ruminococcus albus (strain atcc 27210 / dsm 20455 / jcm 14654 / ncdo 2250 / 7)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2016-03-23 Classification: TRANSFERASE |
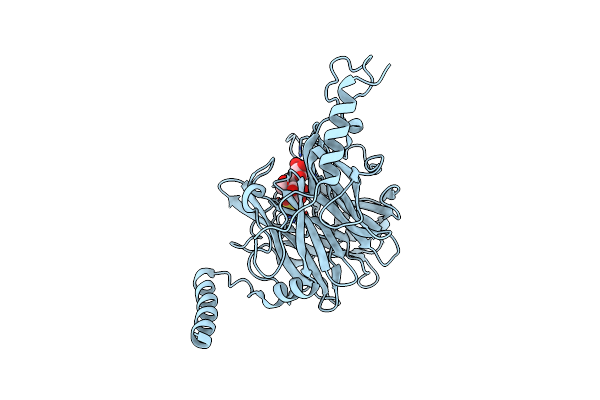 |
Crystal Structure Of Ruminococcus Albus 4-O-Beta-D-Mannosyl-D-Glucose Phosphorylase (Ramp1) In Complexes With Sulfate And 4-O-Beta-D-Mannosyl-D-Glucose
Organism: Ruminococcus albus (strain atcc 27210 / dsm 20455 / jcm 14654 / ncdo 2250 / 7)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-03-23 Classification: TRANSFERASE Ligands: SO4 |
 |
Crystal Structure Of Ruminococcus Albus Beta-(1,4)-Mannooligosaccharide Phosphorylase (Ramp2) In Complexes With Phosphate
Organism: Ruminococcus albus (strain atcc 27210 / dsm 20455 / jcm 14654 / ncdo 2250 / 7)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2016-03-23 Classification: TRANSFERASE Ligands: PO4 |
 |
Crystal Structure Of Ruminococcus Albus Beta-(1,4)-Mannooligosaccharide Phosphorylase (Ramp2) In Complexes With Phosphate And Beta-(1,4)-Mannobiose
Organism: Ruminococcus albus (strain atcc 27210 / dsm 20455 / jcm 14654 / ncdo 2250 / 7)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2016-03-23 Classification: TRANSFERASE Ligands: PO4 |
 |
Crystal Structure Of Pilz Domain Of Cesa From Cellulose Synthesizing Bacterium
Organism: Gluconacetobacter xylinus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2013-04-03 Classification: TRANSFERASE |
 |
Crystallization And Structural Analysis Of Cytochrome C6 From The Diatom Phaeodactylum Tricornutum At 1.5 A Resolution
Organism: Phaeodactylum tricornutum
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2009-03-17 Classification: ELECTRON TRANSPORT Ligands: MG, HEC |
 |
Organism: Pseudomonas sp.
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2001-06-06 Classification: OXIDOREDUCTASE Ligands: FAD, NAD |
 |
Organism: Pseudomonas sp.
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2000-12-20 Classification: OXIDOREDUCTASE Ligands: FAD |

