Search Count: 20
 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Resolution:2.70 Å Release Date: 2024-07-31 Classification: HYDROLASE Ligands: A1L0C |
 |
Organism: Canis lupus familiaris
Method: ELECTRON MICROSCOPY Resolution:2.20 Å Release Date: 2024-07-31 Classification: HYDROLASE Ligands: A1L0D |
 |
Organism: Canis lupus familiaris
Method: ELECTRON MICROSCOPY Resolution:2.00 Å Release Date: 2024-07-31 Classification: HYDROLASE Ligands: A1L0C |
 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Resolution:2.50 Å Release Date: 2024-07-31 Classification: HYDROLASE Ligands: A1L0D, THR |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2021-06-23 Classification: HYDROLASE Ligands: HUR, EDO |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2021-06-23 Classification: HYDROLASE Ligands: HUO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.76 Å Release Date: 2017-12-06 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: RCH |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2016-06-22 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: LWJ, SO4 |
 |
Organism: Camellia sinensis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2014-04-23 Classification: HYDROLASE Ligands: NAG |
 |
Beta-Primeverosidase In Complex With Disaccharide Substrate-Analog N-Beta-Primeverosylamidine, Natural Aglycone Derivative
Organism: Camellia sinensis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-04-23 Classification: HYDROLASE Ligands: VPA, NAG |
 |
Beta-Primeverosidase In Complex With Disaccharide Substrate-Analog N-Beta-Primeverosylamidine, Artificial Aglycone Derivative
Organism: Camellia sinensis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-04-23 Classification: HYDROLASE Ligands: ZPA, NAG |
 |
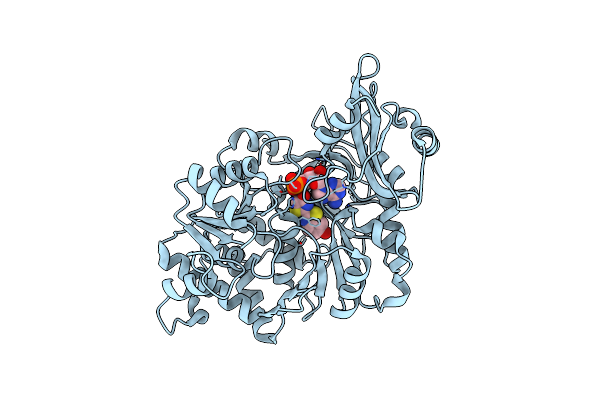
Crystal Structure Of The Thermostable Japanese Firefly Luciferase Complexed With Mgatp
Organism: Luciola cruciata
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2006-03-21 Classification: OXIDOREDUCTASE Ligands: AMP |
 |
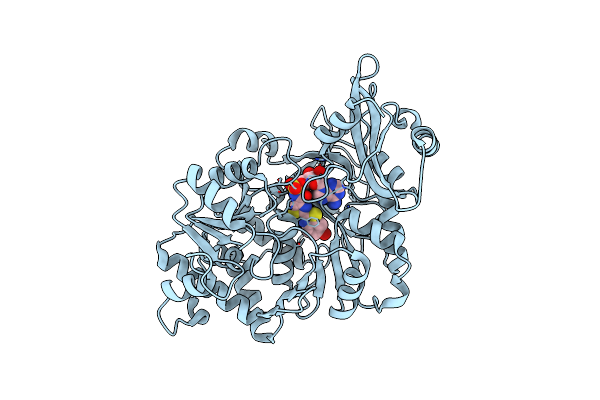
Crystal Structure Of The Thermostable Japanese Firefly Luciferase Complexed With Oxyluciferin And Amp
Organism: Luciola cruciata
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2006-03-21 Classification: OXIDOREDUCTASE Ligands: AMP, OLU |
 |
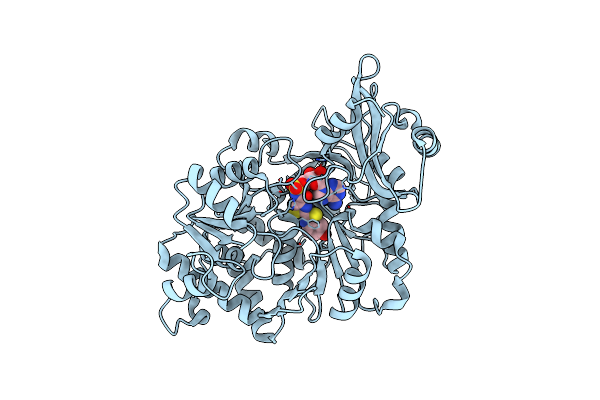
Crystal Structure Of The Thermostable Japanese Firefly Luciferase Complexed With High-Energy Intermediate Analogue
Organism: Luciola cruciata
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2006-03-21 Classification: OXIDOREDUCTASE Ligands: CL, SLU |
 |
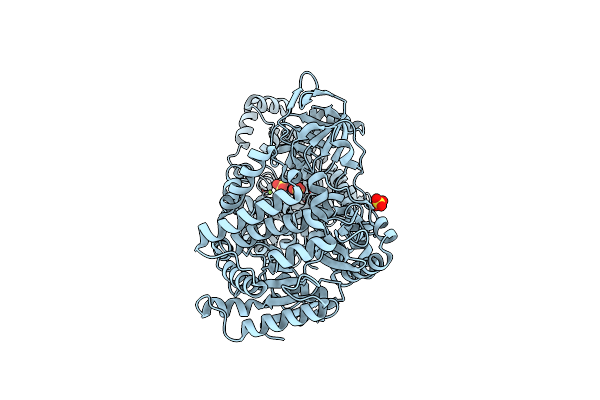
Crystal Structure Of The Thermostable Japanese Firefly Luciferase Red-Color Emission S286N Mutant Complexed With High-Energy Intermediate Analogue
Organism: Luciola cruciata
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2006-03-21 Classification: OXIDOREDUCTASE Ligands: CL, SLU |
 |
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2005-03-08 Classification: TRANSFERASE Ligands: MG, SO4 |
 |
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2005-03-08 Classification: TRANSFERASE Ligands: MG, SO4, PEP |
 |
 |
Crystal Structure Of Candida Albicans N-Myristoyltransferase With Myristoyl-Coa And Peptidic Inhibitor
Organism: Candida albicans
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2002-12-30 Classification: TRANSFERASE Ligands: MYA, MIM |
 |
Crystal Structure Of Candida Albicans N-Myristoyltransferase With Non-Peptidic Inhibitor
Organism: Candida albicans
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2002-12-30 Classification: TRANSFERASE Ligands: R64 |

